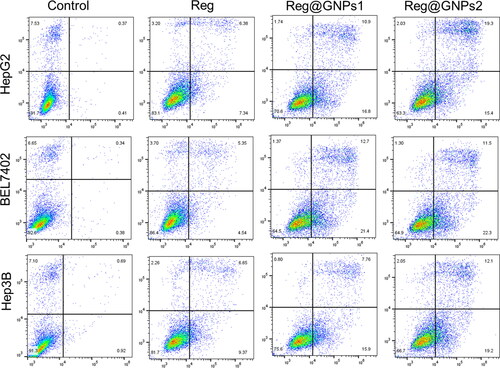
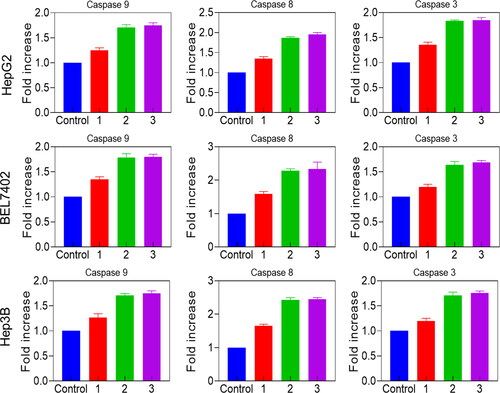
Figures & data
Figure 1. Characterization of GNPs. (A) UV–Vis spectral analysis of Reg@GNPs nanoconjugates dispersions in methanol.
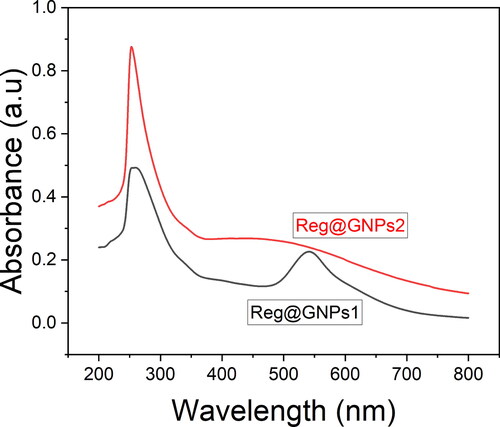
Figure 2. Morphological characterization of GNPs. A and C) TEM images Reg@GNPs1 and Reg@GNPs2, and subsequent SAED patterns. Reg@GNPs1 (upper row) and Reg@GNPs2 (bottom row).
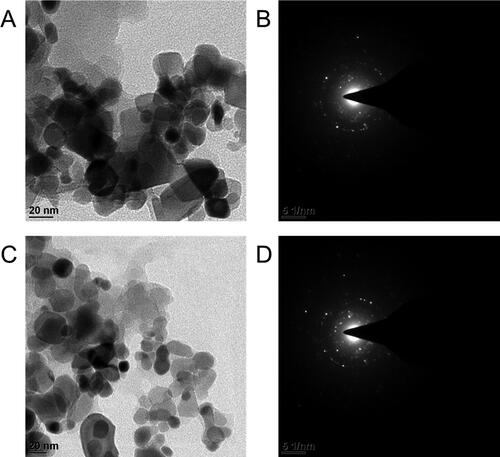
Figure 3. (A) FTIR spectral analysis of Reg and Reg@GNPs nanoconjugates. (B) Power X-ray diffraction spectral analysis of Reg and Reg@GNPs nanoconjugates.
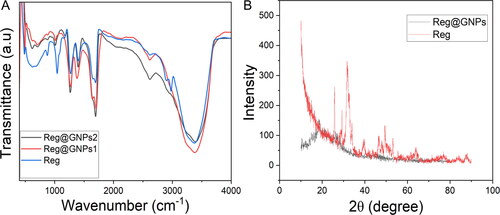
Figure 4. The effect of Reg and Reg@GNPs nanoconjugates on the cell viability of liver cancer cells (Hep3B, BEL7402, and HepG2). cell viability was verified after 24 h of treatment by MTT test. The data are presented as mean ± SD from the representative of three independent experiments.
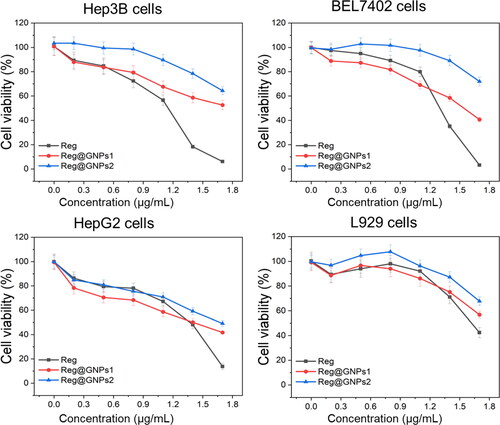
Figure 5. Examination of cell death by acridine orange and ethidium bromide (AO-EB) staining in Hep3B, BEL7402, and HepG2 cells treated with Reg, Reg@GNPs1 and Reg@GNPs2 with their IC50 concentration for 24-h. Scale bar 50 µm.
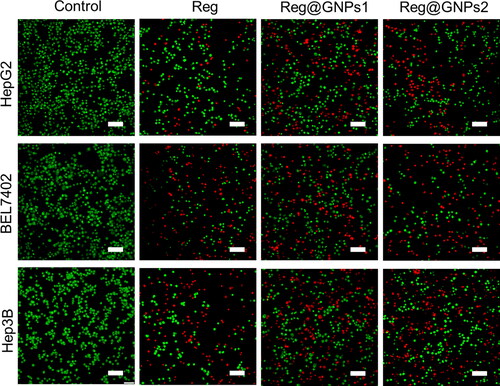
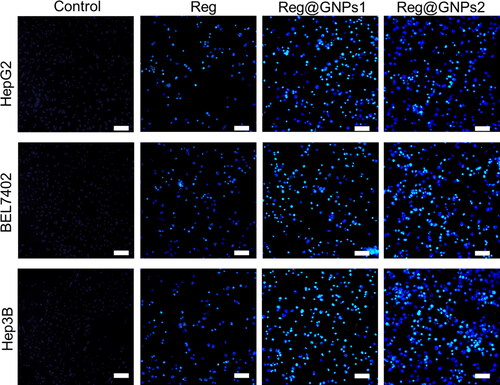
Figure 6. Assessment of nuclear damages by hoechst 33342 staining in Hep3B, BEL7402, and HepG2 cells treated with Reg, Reg@GNPs1 and Reg@GNPs2 with their IC50 concentration for 24-h. Scale bar 50 µm.
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author upon request.