Figures & data
Figure 1. Mobile-phone radiation causes elevation of ROS cellular contents and activation of sporadic cell death, during early and middle oogenesis in Drosophila melanogaster. (A) Confocal laser scanning microscopy (CLSM) images illustrating representative ovarian follicles of stage 10, as they are stained with the general ROS-detector CM-H2DCFDA. Positively reagent-reacted nurse cells were detected only in ovaries of the exposed (EXP) flies (oocyte's green -spotty- signal emanates from auto-fluorescence of yolk granules). (B) Bar-graphs presenting ROS cellular levels ± SEM, as they are measured by suitably used fluorimetric assays, in control (SE: Sham Exposed) and truly exposed (EXP) follicles of all developmental stages from germarium (2b) up to stage 10. (C) Fluorescence microscopy images showing ovarian follicles from sham-exposed (SE) and truly exposed (EXP) flies, as they (follicles) are stained with the acridine orange apoptotic reagent. (D) The ratio of sporadically generated apoptotic follicles was estimated using the number of positively stained, with acridine orange, follicles, of early (germarium 2b) and middle (stages 7–9) oogenesis, against the total number of ovarian follicles of the same developmental stages ± SEM. Scale bars: 50 μm. N = 3, n = 3. SEM: Standard Error Mean. N: number of biological samples; n: number of biological experiments. *: P < 0.05.
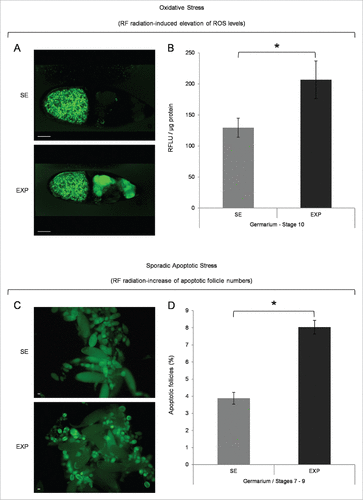
Table 1. Stress-related DEGs upon mobile-phone radiation. Stress-related genes, transcriptionally modified by mobile-phone radiation in Drosophila melanogaster ovarian tissue (>1.25x, P < 0.05). Gene transcription upregulation, or downregulation, is shown (fold and arrows) for the exposed vs. Sham-exposed (control) fly groups.
Figure 2. Mobile-phone radiation alters gene-expression profiling mainly through a transcriptional activation process: a genome-wide approach in fly ovary. Heat-map showing normalized expression levels of 168 identified DEGs (≥1.25 fold, P < 0.05), after application of a microarray-based technology, with their (DEGs) vast majority following an upregulated pattern of expression (compared with control conditions), in response to RF-radiation exposure. Gene-clustering was performed using Euclidean distance and average linkage-analysis software. Light-blue color indicates genes with upregulated levels of expression (153), while gray color specifies genes with downregulated levels of expression (15). SE: Sham Exposed. EXP: Exposed (truly). N = 2, n = 2. N: number of biological samples; n: number of biological experiments. *: P < 0.05.
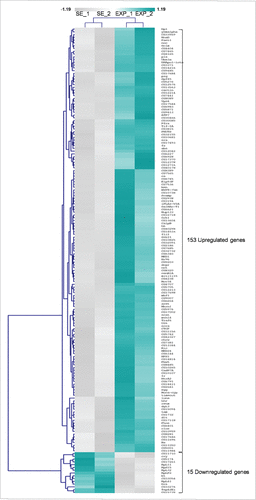
Figure 3. Mobile-phone radiation notably reprograms gene-expression activity in Drosophila melanogaster ovary. (A) Heat-map of DEGs being featured by prominent transcriptional perturbation (≥1.5x, P < 0.05), after RF radiation-induced stress in fly ovary. Gene-clustering was performed using 1.5x cut off, Euclidean distance and average linkage-analysis subroutine. Light-blue color indicates genes with upregulated levels of expression (35), while gray color specifies genes with downregulated levels of expression (4). SE: Sham Exposed. EXP: Exposed (truly). N = 2; n = 2. (B) Validation of gene-microarray data by suitably used RT-qPCR (real time-quantitative polymerase chain reaction) protocols. Bar graph presenting normalized mRNA levels ± SEM. N = 2; n = 3. SEM: Standard Error Mean. N: number of biological samples; n: number of biological experiments. *: P < 0.05.
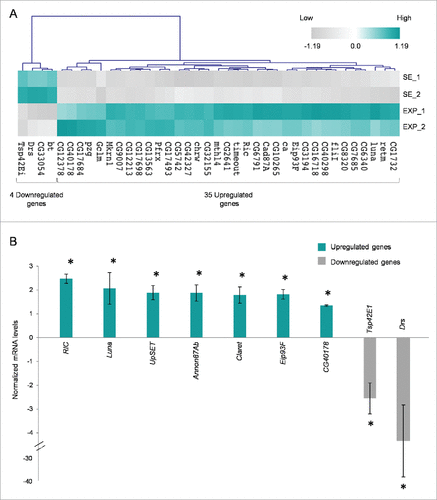
Figure 4. Gene Ontology (GO) and cluster classification analysis of DEGs. Drosophila-ovary genes that have proved to be differentially regulated and expressed by mobile-phone radiation were analyzed using the DAVID-GO bioinformatics subroutine, and they were sequentially categorized according to: (A) “Cellular Compartmentalization” and (B) “General Biological Processes.” (C) Using PANTHER bioinformatics platform, DEG transcripts were also grouped in multiple and diverse “Signaling Pathways.” The number of genes that could be classified in each different category is indicated in parenthesis.
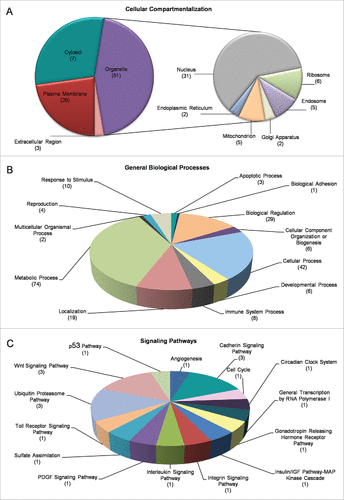
Figure 5. Molecular categorization of DEGs through specific functional annotation. Fly-ovary genes shown to be differentially controlled and expressed in response to mobile-phone radiation exposure were processed through engagement of DAVID-GO bioinformatics tool and they were further classified according to their: (A) “Specific Binding Interactions” and (B) “Specific Catalytic Activities.” The number of genes that could be embraced by each different category is indicated in parenthesis.
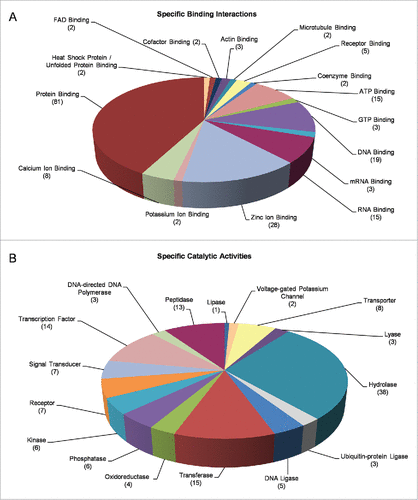
Table 2. Cell-death-related DEGs upon mobile-phone radiation. Programmed cell death-related genes, transcriptionally activated by mobile-phone radiation in Drosophila melanogaster ovarian tissue (>1.25x, P < 0.05). Gene transcription upregulation is denoted, for the exposed vs. control fly groups, in the form of fold and arrows.
Figure 6. Figure 6. Cataloguing and functional clustering of DEG human orthologs through engagement of Ingenuity Pathway Analysis (IPA) tool. (A) Bar-graphs denoting fly-ovary DEGs with statistically significant (P < 0.05) homologies to human counterparts and their cognate reconstructed protein pathways. Ratio is referred to the number of genes from our DEG list [comparison of their transcriptional activities between SE (sham exposed) and EXP (truly exposed) fly groups] divided by the total number of genes involved in each specific pathway that has been notably affected by flies' exposure to mobile-phone radiation. Threshold is determined at P value 0.05 [-log (0.05) = 1.30]. To the same direction, RF-radiation exposure proved able to differentially perturb the transcriptional-expression profiling of several genes (DEG human orthologs) being critically implicated in: (B) “Cell Cycle” - “Cell Signaling,” (C) “Cell Death” - “Cell Survival,” (D) “DNA Replication” - “DNA Recombination” - “DNA Repair” and (E) “Embryonic Development,” as clearly revealed through suitable application of IPA bioinformatics tool. Red color indicates the upregulated gene products, while green color specifies the downregulated ones. Figures were automatically produced via IPA-software protocols.
![Figure 6. Figure 6. Cataloguing and functional clustering of DEG human orthologs through engagement of Ingenuity Pathway Analysis (IPA) tool. (A) Bar-graphs denoting fly-ovary DEGs with statistically significant (P < 0.05) homologies to human counterparts and their cognate reconstructed protein pathways. Ratio is referred to the number of genes from our DEG list [comparison of their transcriptional activities between SE (sham exposed) and EXP (truly exposed) fly groups] divided by the total number of genes involved in each specific pathway that has been notably affected by flies' exposure to mobile-phone radiation. Threshold is determined at P value 0.05 [-log (0.05) = 1.30]. To the same direction, RF-radiation exposure proved able to differentially perturb the transcriptional-expression profiling of several genes (DEG human orthologs) being critically implicated in: (B) “Cell Cycle” - “Cell Signaling,” (C) “Cell Death” - “Cell Survival,” (D) “DNA Replication” - “DNA Recombination” - “DNA Repair” and (E) “Embryonic Development,” as clearly revealed through suitable application of IPA bioinformatics tool. Red color indicates the upregulated gene products, while green color specifies the downregulated ones. Figures were automatically produced via IPA-software protocols.](/cms/asset/c9ed0e00-2c6a-45ff-a844-28e25fb28da0/kfly_a_1270487_f0006_oc.gif)
Table 3. DEGs with putative binding sites of ROS-dependent transcription factors. Catalog of fly-ovary DEGs being characterized by the in silico presence (P < 10−4) of the, herein, denoted transcription-factor (AP-1, FOXO3, FOXO4, HIF-1α, NRF2, p53 and REL/NFκB) binding sites in their (DEGs) (semi-)proximal -respective- promoter regions [-1 to −500 bp, upstream (5′) from the transcription start site (TSS: +1)]. Each one of the, herein, examined transcription factors has been previously reported to critically control redox homeostasis under certain cellular settings and stress conditions [41–45]. Identification of binding sites was performed with FIMO algorithm from MEME suite (http://meme-suite.org/) [46] with default settings, using the motifs identified from JASPAR database (http://jaspar.genereg.net), a high-quality transcription factor-binding profile database. The number of times (x) a binding site is recognized in an examined-gene (DEG) promoter is described in its respective parenthesis (e.g., 2x or 3x), when required to be given.
