Figures & data
Figure 1. A Nedd8 mutation designed to prevent deneddylation. (A) The COP9 signalosome deneddylates Cullins, reducing the activity of CRL ubiquitin ligase complexes. We predicted that uncleavable Nedd8 would have the same effect as CSN loss of function, preventing Cullin deneddylation and increasing CRL activity. (B) Diagram of the UAS-Nedd8 transgenes that were either transfected into S2 cells or injected into Drosophila embryos. Uncleavable Nedd8 harbors an amino acid change from leucine to proline at position 73.
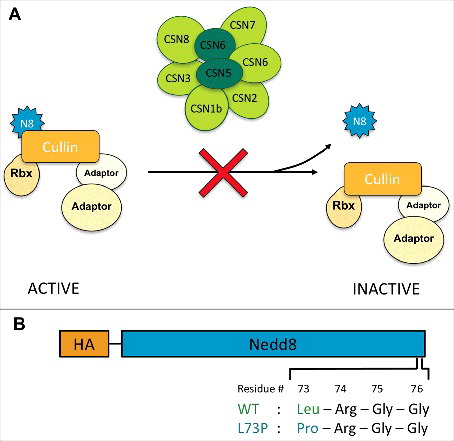
Figure 2. Nedd8L73P is conjugated onto Cullins and does not dominantly affect development. (A) Lysates from S2 cells transfected with FLAG-Cul3 alone or together with HA-Nedd8WT or HA-Nedd8L73P were immunoprecipitated with anti-HA and blotted with anti-FLAG (upper panel). Lower panels show the input blotted with anti-FLAG, anti-HA, anti-Arm and anti-β-tubulin. More Nedd8L73P than wild-type Nedd8 is present as a Cul3 conjugate. (B) Lysates of wing discs in which HA-Nedd8WT or HA-Nedd8L73P was specifically expressed only in GFP-labeled Nedd8 mutant clones, blotted with anti-HA, anti-Nedd8, anti-GFP and anti- β-tubulin. Both forms of Nedd8 are present in a band at the expected molecular weight of Drosophila Cullins, but Nedd8L73P gives rise to stronger HA and Nedd8 labeling of this band. (C-E) Wing discs stained with anti-HA (C’, D’, E’, green in C, D, blue in E) and anti-Ci (C’’, D’’, E’’, magenta in C, D, red in E). HA-Nedd8WT (C) or HA-Nedd8L73P (D) is driven in the dorsal compartment by ap-GAL4, but has no dominant effect on Ci expression. In (E), HA-Nedd8WT is driven with ap-GAL4 in a wing disc in which Nedd8 mutant clones are marked by the absence of RFP (green). Transgenic tagged Nedd8 rescues Ci accumulation and clone growth in the dorsal compartment, and accumulates to higher levels in clones that lack endogenous Nedd8.
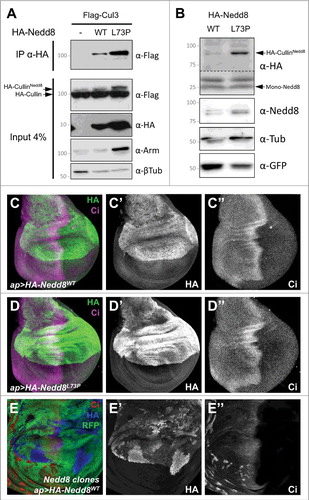
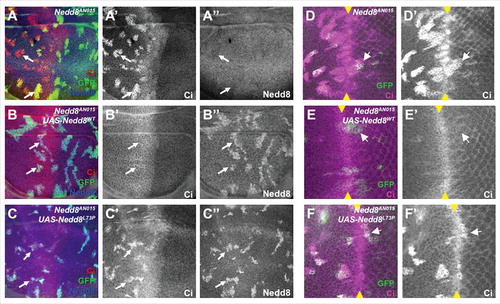
Figure 3. Nedd8L73P does not rescue CRL function. (A, B, C) Wing discs and (D, E, F) eye discs in which Nedd8 mutant clones are positively labeled with GFP (green) and express no transgene (A, D), HA-Nedd8WT (B, E), or HA-Nedd8L73P (C, F). Discs are stained with anti-Ci (A’, B’, C’, D’, E’, F’, red in A, B, C, magenta in D, E, F) and anti-Nedd8 (A’’, B’’, C’’, blue in A-C). Ci, which is a substrate for a Cul1 ligase in the wing disc and anterior eye disc and for a Cul3 ligase in the posterior eye disc, accumulates in Nedd8 mutant clones and Nedd8 mutant clones rescued with Nedd8L73P, but not in Nedd8 clones rescued with Nedd8WT. Arrows indicate representative clones, and yellow arrowheads mark the morphogenetic furrow in the eye discs.