Figures & data
Figure 1. Anti-VLP peptides selected from phage-displayed peptide libraries. (A) The sequences of unique clones, denoted by the single-letter amino acid code, selected against WT, D66H or L55F are listed separately. The parental clones, Pep-1 and Pep-2, isolated against WT, are listed at the top. Cysteine residues are shaded yellow. The relative binding strength of each clone to different VLPs was qualified according to their phage ELISA signals against different VLPs, −: <0.5; +: 0.5–1; ++: 1–1.5; +++: 1.5–2; ++++: 2–2.5; +++++: >2.5. (B) Binding of peptide-Fc fusion proteins against a panel of VLP variants. ELISA signals are presented as mean values ± SD from 2 independent measurements.
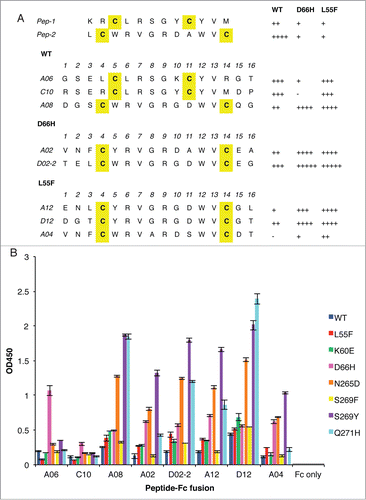
Figure 2. CDR sequences of anti-L55F antibodies. CDR-L3 and the 3 heavy chain CDRs are shown, as CDR-L1 and CDR-L2 were not diversified in the library design. The numbering is according to the nomenclature of Kabat et al.Citation39 Residues in specificity-tuned clones that are different from the parental clone GC058 are shown in white with black background. The signal ratios of Fab-phage bound to L55F vs. WT in phage ELISA (L55/WT) were determined as the mean values from 3 independent measurements.
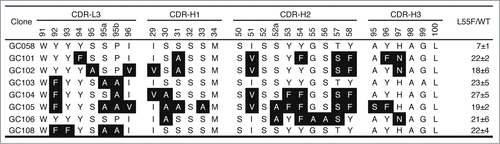
Figure 3. Positional amino acid preferences in Fab variants from homolog scanning of GC058 for functional selection against L55F (A) or display selection against anti-FLAG antibody M2 (B). For each position, the percentage of the population is shown for the wild type or its homolog residue. The wild type GC058 residue is indicated prior to position number, and its homolog residue following the number. The data were compiled from 39 or 40 unique sequences from the functional selection or the display selection, respectively.
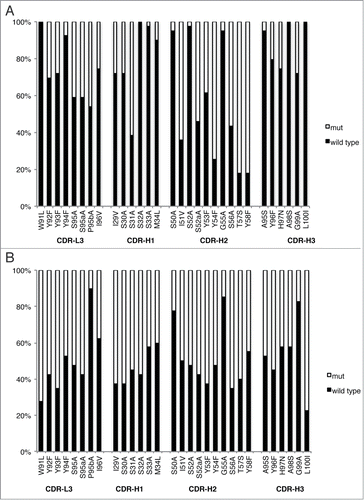
Figure 4. Detection of WT and L55F VLPs by Fabs. ELISAs were performed with serial dilutions of Fab proteins using plates coated with 1 μg/ml of WT VLPs (A) or L55F VLPs (B). Data from L55F-binding ELISAs were fit to a 4-parameter logistic equation.
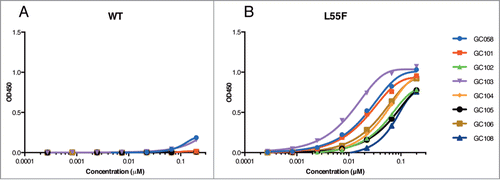
Figure 5. Detection of VLP variants by Fabs. ELISAs were performed with 10 μg/ml of Fab proteins using plates coated with 1 μg/ml of the indicated VLPs. ELISA signals are presented as mean values ± SD from 2 independent measurements.
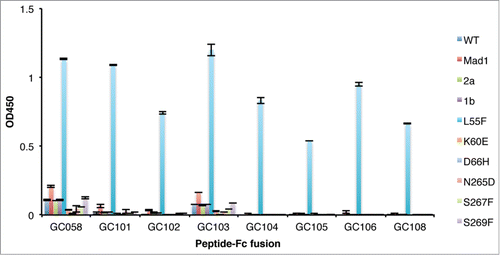
Figure 6. Detection of VLPs by IgGs. ELISAs were performed with serial dilutions of IgG proteins using plates coated with 1 μg/ml of WT VLPs (A), L55F VLPs (B) or D66H VLPs (C). ELISA signals are presented as mean values ± SD from 3 independent measurements.
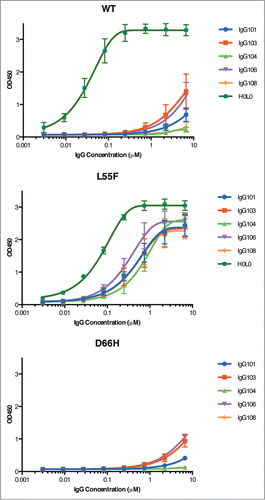
Table 1. Mutagenic oligonucleotides used for peptide and antibody library constructionFootnote*
