Figures & data
Figure 1. The native-like FcRn binding could be re-built by transplanting a short FcRn binding motif of IgG1 CH3 onto IgG1 CH2 domain. (a) The complex structure of rat FcRn with Fc (PDB entry 1FRT). The Fc CH2 domain was colored in orange and the Fc CH3 domain was colored in cyan. (b) A schematic diagram to describe the in silico design of CH2-CH3 hybrids. One chain of IgG1 Fc dimer compromising one CH2 and one CH3 domain was shown, and the FcRn binding motif in the CH3 domain was colored red. The CH2 domain and the FcRn binding motif of CH3 were joined by a randomized peptide loop. The residues considered to be important for FcRn binding (H433, H435 and Y436) in CH3 were highlighted.
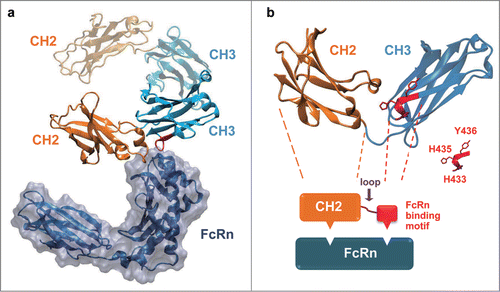
Figure 2. Schematic representation of the engineering strategy to generate pH-dependent FcRn binders. (a) The overlapping view of 6 modeling structures of CH2-CH3 hybrids that possess randomly selected peptide linkers between m01s and FcRn binding motif of CH3. The molecule with a 4-mer linker was colored red, with a 6-mer linker was colored orange while all others have 8-mer linkers. Two histidines in the CH3 motif were drawn as bonds, showing different position and orientation of this motif with relative to m01s. (b) schematic representation of the panning method for the enrichment of only pH-dependent FcRn binders, as described in Experimental Procedures. (c) Plots of the change in fraction folded (calculated from CD molar ellipticity at 216 nm) for the isolated CH2 domain, 2C10, a2b and m01s.
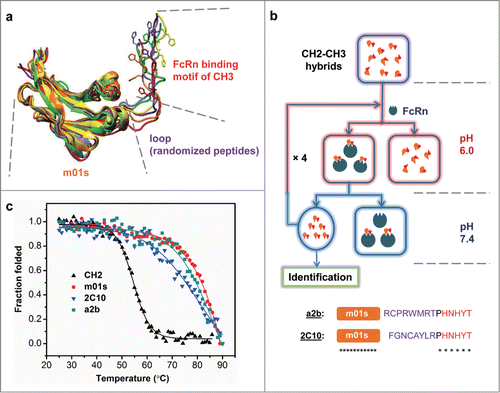
Figure 3. Measurement of aggregation for IgG1 CH2, m01s, 2C10 and a2b by dynamic light scattering with the X-axis denoting protein particle size (diameter, nanometer) and the Y-axis intensity (%). Three different colors represent 3 measurements of the same protein.
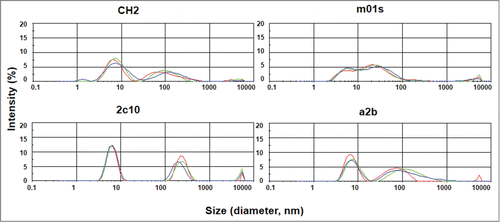
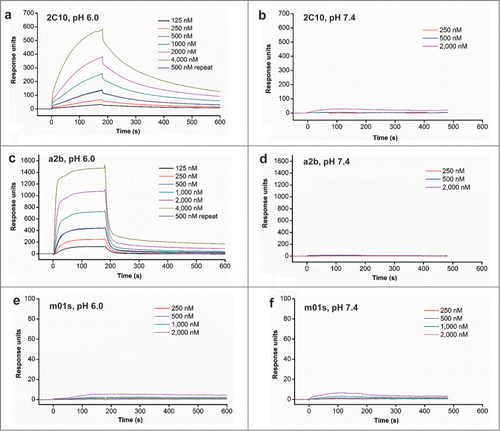
Figure 5. FcRn binding of measured by flow cytometry and ELISA. (a) Binding of MDCK-FcRn cells to 2C10, a2b, m01s and wild-type Fc (1 µM) at pH 6.0 (left) and 7.4 (right) analyzed by flow cytometry. (b) Schematic representation of the constructs with the FcRn binding motif or the conformational linker removed from 2C10 or a2b. (c) Binding of different antibody domain constructs to human single-chain soluble FcRn measured by ELISA at pH 6.0.
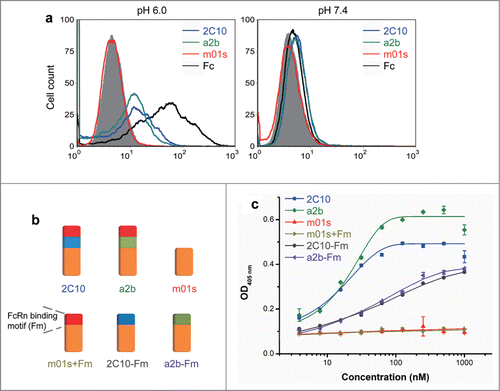
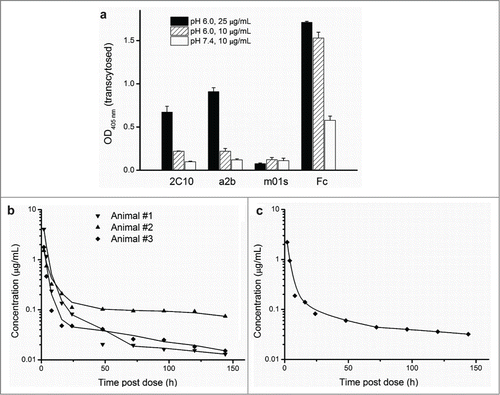
Figure 6. The improved FcRn binding correlates with significantly enhanced transcytosis and prolonged in vivo half-life. (a) In vitro transepithelial transport of m01s, a2b, 2C10 and Fc at 10, 25 ug/mL (pH 6.0) and 10 ug/mL (pH 7.4). (b) Pharmacokinetics of 2C10 dosed to individual primates numbered 1, 2 and 3. (c) Serum concentration-time course plot of pooled serum samples of the same 3 animals.