Figures & data
Figure 1. Amino acid sequence alignment of the anti-TetC variable domains. The deduced amino acid sequences are given in single-letter code. Residues are numbered according to the IMGT numbering system. Dots are used to denote identical residues and dashes to indicate gaps. Dotted boxes frame the CDRs and solid-line boxes outline the characteristic hallmark residues that differ between VHs and VHHs.

Figure 2. Epitope mapping and in vitro inhibition of TetC binding to GT1b. (a) Epitope binning and affinity measurement to TetC by bio-layer interferometry: The vertical dotted lines separate the steps (30 s) in the sequential exposure of nanobodies to the sensor. (b) Inhibition of GT1b binding of TetC coupled to peroxidase with the 3 VH/VHHs; VHH-TC9 against triclocarbanCitation29 was used as a negative control, all measurements are triplicates. (c) Schematic representation of the VHH epitopes according to the binning analysis and GT1b binding inhibition. The GT1b binding site is represented by the dashed area. The error bars represent the SD of mean value.
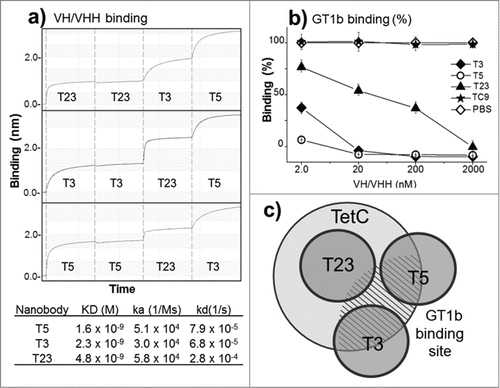
Figure 3. In vivo neutralization of high (a) and low (b) doses of toxin with monomeric antibody fragments. Groups of 5 mice were co-administered with: (a) 10 × LD100 of the toxin preparation and 6.3 nmoles of T5, or 3.3 nmoles of Poly-IgG, or PBS ; or (b) 1 × LD100 of the toxin preparation and 0.63 nmoles of T5, 0.63 nmoles of T3, 0.33 nmoles of Poly-IgG, or PBS.
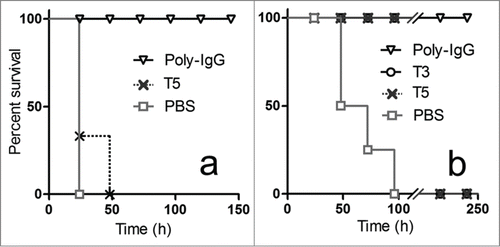
Figure 4. Expression of the bivalent nanobody T5–T3 and in vivo neutralization assay. (a) SDS-PAGE 12% analysis of the cell culture extract of VH-VHH T5–T3 (1) or VH T5 (2), and the affinity purified T5–T3 (3). (b) Survival rate of groups of 5 mice co-administered with 10 × LD100 of the toxin preparation and PBS or PBS containing 6.3 nmoles of T5, 6.3 nmoles of T5–T3, or 3.3 nmoles of Poly-IgG.
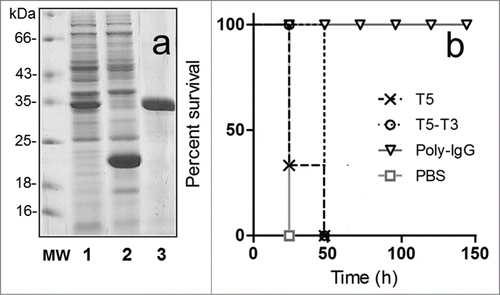
Figure 5. Comparison of the reactivity of the monomeric and bispecific nanobodies. (a) Analysis of the nanobody reactivity against TetC by ELISA and bio-layer interferometry. (b) Flow cytometry mean fluorescent intensity (MFI) of the J774.A1 cell line stained with the biotinylated monomeric or bispecific nanobodies and streptavidin phycoerythrin. The insert shows the histogram for the 1 nM concentration of V36 (black), V36-T5 (dark gray), T5–V36 (light gray), and TC9 (negative control, dashed line). The error bars represent the SD of mean value.
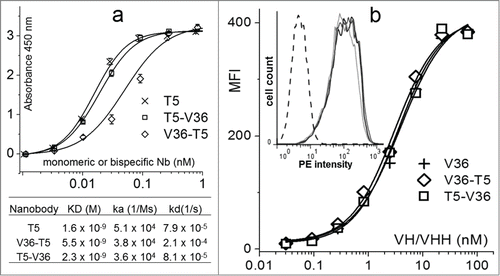
Figure 6. In vivo neutralization assay with bispecific antibodies. Survival rate of groups of 5 mice co-administered with 10 × LD100 of the toxin preparation and 6.3 nmoles of the monomeric or bispecific antibodies in PBS. The Log-rank (Mantel-Cox) test indicated significant differences in the neutralizing activity of the nanobodies as follows: T5 < T5 – TC9 (p < 0.0002), T5 – TC9 < T5 – 49 (p < 0.0002), T5 – 49 < T5 – G7(p<0.02), T5 – G7 <T5 – V36 (p < 0.0001).
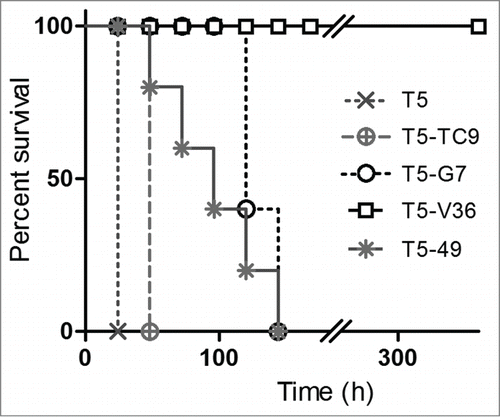
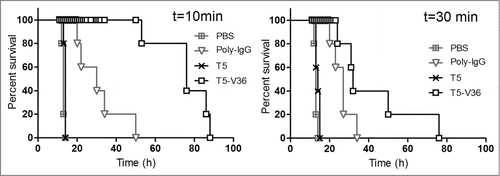
Figure 7. Delayed neutralization assay. Survival rate of groups of 5 mice were challenged s.c. with 5 × LD100 of the toxin preparation, and 10 and 30 min later, treated i.v. with PBS, or PBS containing 6.3 nmoles of T5, T5–V36, or 3.3 nmoles of Poly-IgG. The Log-rank (Mantel-Cox) test indicated significant differences in the neutralizing activity of the nanobodies as follows: PBS<Poly-IgG (p < 0.0001), Poly-IgG <T5 – V36 (p < 0.0001) at 10 min, and PBS <Poly-IgG (p < 0.0001), Poly-IgG <T5 – V36 (p < 0.05) at 30 min.