Figures & data
Figure 1. Representative total ion chromatogram of the BSA digest spiked with synthetic peptides used in this study for evaluation of system suitability. The intra-scan peptides (highlighted in red) were spiked in pairs consisting of one light peptide and one heavy isotope labeled peptide (schematic mass spectrum representation shown in insert). The light peptides were present at signal normalized concentrations corresponding to 0.1% to 100% of the heavy peptide concentrations in each pair. The inter-scan peptides (highlighted in blue) are closely related synthetic peptides eluting at similar retention times around 30 min. The other peaks in the chromatogram correspond to BSA peptides. The signal from the inter-scan peptides present at signal normalized concentrations of 1% and 0.1% are too low to be directly observed on the total ion chromatogram.
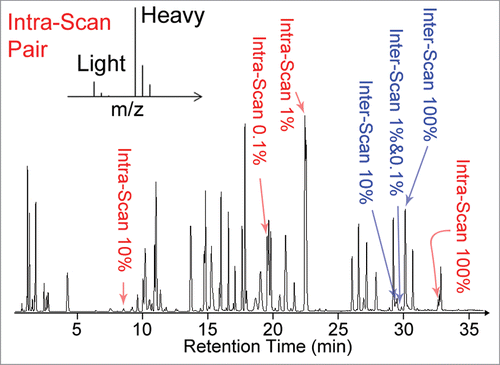
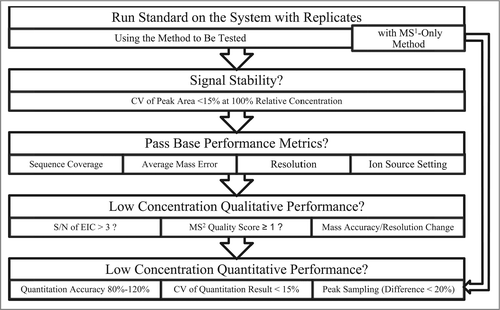
Table 1. Experimental method settings with systematic variation of several parameters. “+” sign indicates high source voltage; long MS1/MS2 scan time/accumulation; or high MS2 precursor selection threshold. “−” sign corresponds to lower settings. M0 is a MS1 only method, thus the MS2 parameters are listed as “NA” (not applicable)
Figure 2. Influence of method settings on protein sequence coverage (black bars) for data from (a) Q-Exactive and (b) QTOF instrument as reported by Proteome Discoverer. Gray bars indicate the average percentage of MS2 spectra matched to peptides for each method (number of spectra associated with high-confidence peptides divided by the total number of spectra input into the search algorithm). The number of MS2 spectra inputs into the search algorithm is also included as line plots following the y-axis on the right.
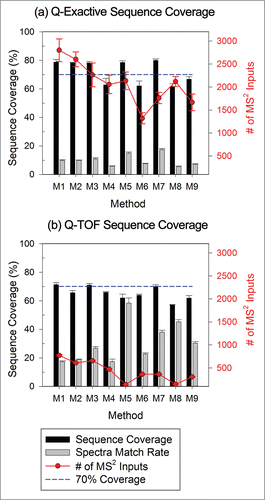
Figure 3. MS2 scores for inter-scan peptide standards at signal normalized concentrations of 10% for Q-Exactive and QTOF systems. Higher scores were associated with higher confidence in identification. Diamonds indicate the confidence interval, with the center line indicating the mean and the vertical span representing the 95% confidence interval.
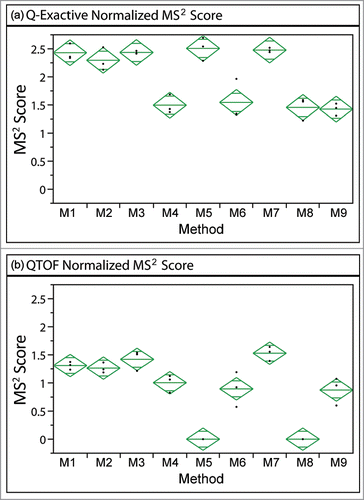
Figure 4. Average absolute mass error associated with each acquisition method on either MS platform. ANOVA analysis identifies significant differences among the Q-Exactive methods (P< 0.0001), but not for the QTOF methods (p = 0.2097). A pairwise t-test, however, suggests that significant differences in mass error between some of the methods on QTOF exist.
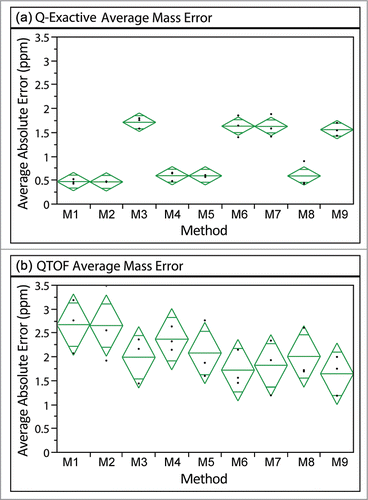
Figure 5. Average absolute ppm errors of high-confidence BSA peptides identified for data acquired on the Q-Exactive and QTOF sorted by differences in MS1 scan time. The (+) corresponds to a higher MS1 AGC target (Q-Exactive) or a longer MS1 scan (QTOF) than the (−). Significant differences were observed between the different MS1 settings on both systems (p < 0.0001 for Q-Exactive and p = 0.0083 for QTOF).
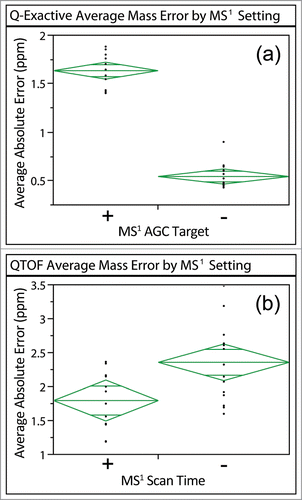
Figure 6. Summary of accuracy and precision results for intra-scan (left column) and inter-scan (right column) peptide pairs at different signal normalized concentration ratios on the Q-Exactive (top row) and QTOF (bottom row). Percent accuracy is defined as the experimental result divided by the expected value. The precision is represented by the standard deviation as shown with the error bars. Missing points on broken lines indicate the peptide was not detected at least twice in the triplicate runs using the associated method. Data with CV > 20% are highlighted with yellow edges on the points.
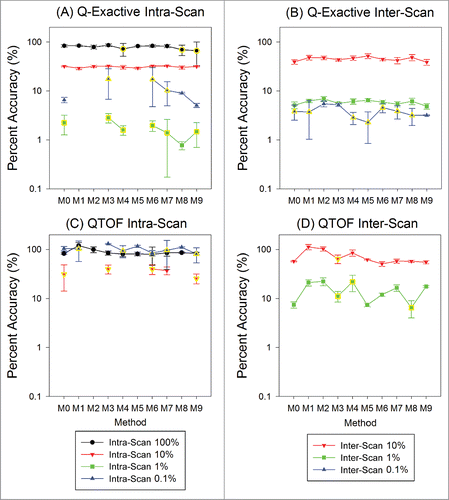
Figure 7. Variation in quantitation results relative to the MS1 only method (M0) due to variation in peak sampling for 2 inter-scan peptides on both the QTOF and Q-Exactive. The sampling factor featured good correlation with the QTOF data, but not with the Q-Exactive data as manifested by the correlation coefficients (R) noted in the plots. The ranges where the normalized results are 0.8∼1.2 (relative quantitation result within ± 20% of that by method M0) are highlighted in blue in each plot.
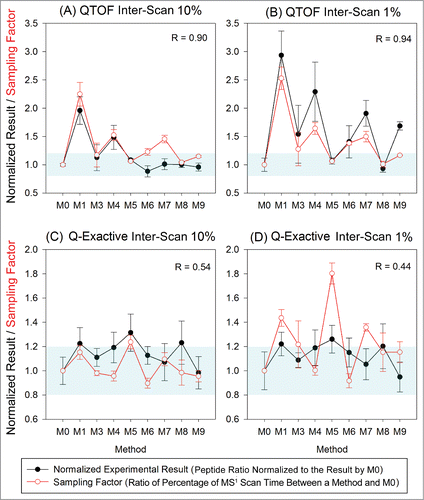
Table 2. Summary of the proposed metrics for system suitability test, with proposed acceptance criteria and possible troubleshooting targets. The thresholds of 15% for CV and +/− 20% for relative quantitation are adapted from current FDA recommendationsCitation34