Figures & data
Figure 1. Structures of the plasmids used to generate transgenic silkworms. Each plasmid has right and left arms of piggyBac and the 3 × P3-fluorescent gene cassette for a screening marker (EYFP, AmCyan, or DsRed2). Plasmids pBac[UAS_anti-CD20 mAb HC/3 × P3-EYFP] and pBac[UAS_anti-CD20 mAb LC/3 × P3-AmCyan] encode the anti-CD20 mAb H chain gene and the anti-CD20 mAb L chain gene, respectively, under the control of a UAS promoter, and contain an BmNPV- derived hr5 enhancer and an A3-Blasticidin cassette. The anti-CD20 H and L genes were fused to the signal peptide sequence of the sericin 1 gene encoded by exon 1 and 18 bp of exon 2. The plasmid pBac[Ser1-GAL4/3 × P3-DsRed] encodes the GAL4 gene under the control of the sericin1 promoter.
![Figure 1. Structures of the plasmids used to generate transgenic silkworms. Each plasmid has right and left arms of piggyBac and the 3 × P3-fluorescent gene cassette for a screening marker (EYFP, AmCyan, or DsRed2). Plasmids pBac[UAS_anti-CD20 mAb HC/3 × P3-EYFP] and pBac[UAS_anti-CD20 mAb LC/3 × P3-AmCyan] encode the anti-CD20 mAb H chain gene and the anti-CD20 mAb L chain gene, respectively, under the control of a UAS promoter, and contain an BmNPV- derived hr5 enhancer and an A3-Blasticidin cassette. The anti-CD20 H and L genes were fused to the signal peptide sequence of the sericin 1 gene encoded by exon 1 and 18 bp of exon 2. The plasmid pBac[Ser1-GAL4/3 × P3-DsRed] encodes the GAL4 gene under the control of the sericin1 promoter.](/cms/asset/a6a011b9-193b-42d9-9b12-b88525fe552e/kmab_a_1078054_f0001_oc.gif)
Figure 2. Expression of anti-CD20 mAb in transgenic silkworms. (A) The protein lysates extracted from MSGs of H line or L line transgenic silkworms were separated by SDS-PAGE followed by staining with CBB or by western blotting with an anti-Human IgG(H+L) antibody. The protein lysates extracted form MSGs of transgenic silkworms that harbored only the Ser1-GAL4 construct were used as negative controls. The numbers above the gels or western blots indicate the line number of each transgenic strain. (B) The protein lysates extracted form MSGs or cocoons of the H+L line transgenic silkworms were separated by SDS-PAGE under reducing or non-reducing conditions. Gels were stained with CBB or subjected to protein gel blotting using an anti-Human IgG(H+L) antibody. The lysates obtained from the transgenic silkworms harboring only the Ser1-GAL4 construct were used as a negative control.
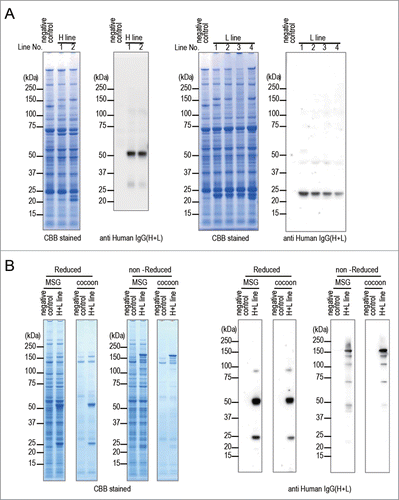
Figure 3. Purification of anti-CD20 mAbs from the transgenic silkworms. (A) Anti-CD20 mAbs were purified from the lysates of MSGs or cocoons derived from transgenic silkworms by affinity chromatography using a Protein G column. The lysates (original), flow-through fractions from the Protein G column, and eluted fractions were analyzed by non-reducing SDS-PAGE. (B) MabThera and anti-CD20 mAbs derived from transgenic silkworms showed the same migration pattern on non-reducing SDS-PAGE.
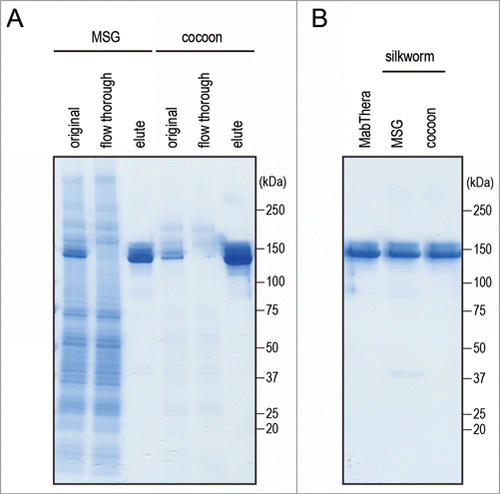
Figure 4. CD20-binding properties. (A) Daudi cells were incubated with anti-CD20 mAbs and their binding properties were assessed by flow cytometric analysis using Alexa488-conjugated F(ab')2 anti-human IgG Fc. The mean fluorescence intensities were plotted against the mAbs concentration (n = 3, bars indicate SEM). (B) Daudi cells were incubated with DyLight 488-labeled MabThera and serially diluted unlabeled mAbs. Percentages of DyLight 488-labeled MabThera binding were calculated by the mean fluorescence intensities and plotted against the unlabeled mAbs concentration (n = 3, bars indicate SEM).
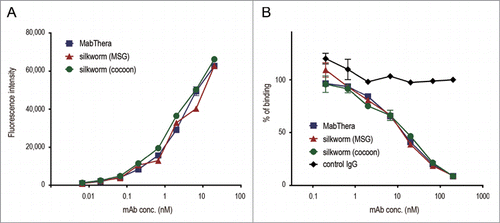
Figure 5. Binding affinity to human Fc receptors. Binding affinities of anti-CD20 mAbs to FcγRI, FcγRIIIa and FcRn were measured by an SPR analysis. (A) Binding sensorgrams corrected for both the surface blank and the buffer injection control are represented. Dashed lines indicate the fitting curves generated by 1:1 binding model for FcγRs and bivalent model for FcRn, respectively. (B) Dissociation constant (KD) values are represented as mean + SD (n = 3). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
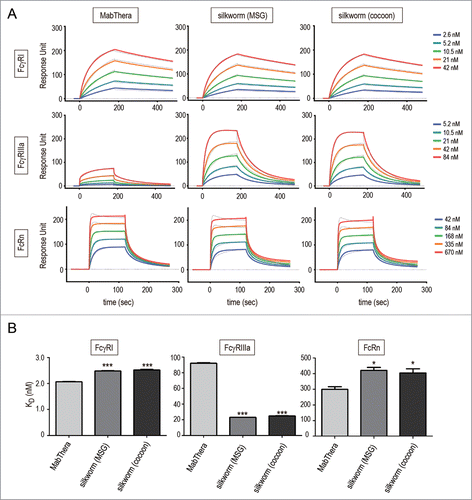
Table 2. Primers used in this study
Table 1. Binding affinities to human Fc receptors
Figure 6. FcγRIIIa activation and antibody-dependent cell-mediated cytotoxicity (ADCC). (A) CD20 binding-dependent activation of FcγRIIIa was measured by using Daudi (target) and Jurkat/FcγRIIIa/NFAT-Luc (effector) cells. Fold increases in luciferase activity were plotted against the mAbs concentration (n = 3, bars indicate SEM). (B) The levels of ADCC against Daudi cells were measured by using human PBMCs as effector cells. The percentages of cell lysis were plotted against the mAbs concentration (n = 3, bars indicate SEM).
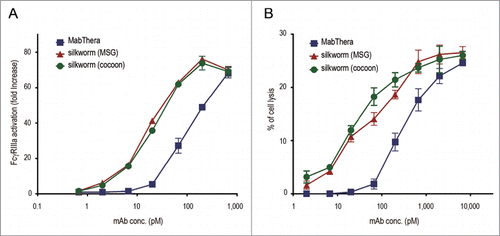
Figure 7. Complement-dependent cytotoxicity (CDC). Daudi cells were cultured in the presence of human serum (16%) and anti-CD20 mAbs (1, 3 or 10 µg/ml). The percentages of 7-AAD positive-dead cells were calculated by flow cytometric analysis and represented as the mean + SEM (n = 3). **, p < 0.01; ***, p < 0.001.
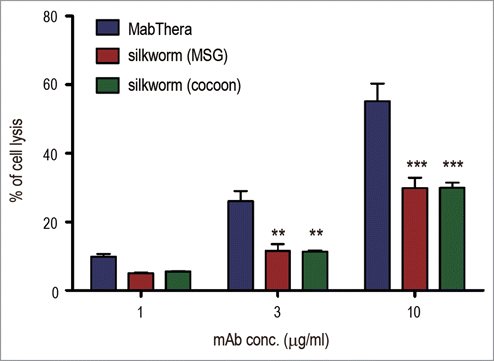
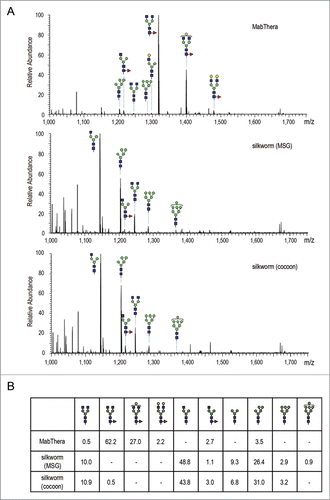
Figure 8. Fc glycosylation analysis of anti-CD20 mAbs. (A) Comparison of mass spectra acquired at the elution point of the glycopeptides of EEQYNSTYR, corresponding to 293–301 amino acids (EU numbering) in the heavy chain of anti-CD20 mAbs. (B) Percentage distributions of glycoforms were calculated by the relative peak area average values. Symbols: blue square, N-acetylglucosamine; green circle, mannose; yellow circle, galactose; red triangle, fucose.