Figures & data
Figure 1: (a) Schematic representation of the in vitro affinity maturation steps applied in the selection of high affinity GM-CSF antibodies. KD values are indicated. Citation17 (b) Sequence overview of CDR-H2 and CDR-L3 of anti-GM-CSF antibodies MOR03929, MOR04302, MOR04252 and MOR04357. Kabat Citation27 numbering scheme is shown.
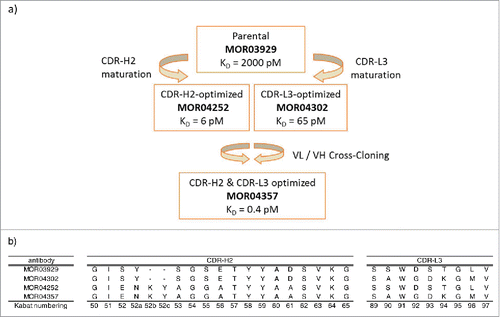
Figure 2. Overall structure and superposition of all 4 Fab:GM-CSF complexes. (a) Structure of MOR04357:GM-CSF as cartoon presentation. Helices are numbered as letters A-D and β-sheet as numbers 1 and 2. N- and C-termini are depicted as N and C. MOR04357 Ig light chain is colored in green, the Ig heavy chain in blue. CDR regions are shown as L1-L3 and H1-H3 with respect to the corresponding chain. (b) GM-CSF and the variable domain of Fab fragment as ribbon presentation. Complex of MOR03929:GM-CSF is presented in yellow, MOR04252:GM-CSF in green, MOR04302:GM-CSF in blue and MOR04357:GM-CSF in red.
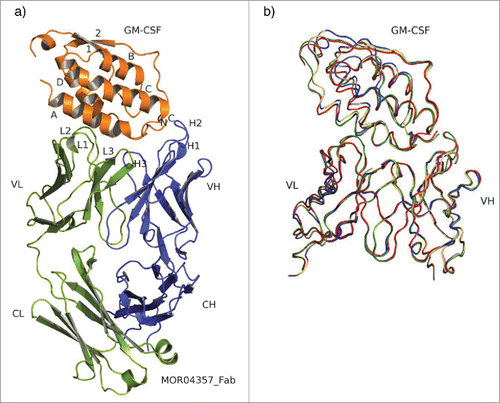
Figure 3. Cartoon representation of free MOR04357 (pale colors) and MOR04357 in complex with GM-CSF (dark colors). GM-CSF is presented in orange, the Ig light chain in green colors and the Ig heavy chain in blue colors. Respective CDR-L2 loops are presented in yellow and light yellow. Distances are given in angstrom and shown in red, angles are given in degree [°] and presented in black. Val-47 of both structures are shown as stick presentation. Val-47 undergoes a backbone flip from −64.5° to 154.4° when MOR04357 is bound to GM-CSF compared to the uncomplexed MOR04357. This backbone flip results in a shift of 8.2 Å of this loop toward GM-CSF along with a rotation by 39°.
![Figure 3. Cartoon representation of free MOR04357 (pale colors) and MOR04357 in complex with GM-CSF (dark colors). GM-CSF is presented in orange, the Ig light chain in green colors and the Ig heavy chain in blue colors. Respective CDR-L2 loops are presented in yellow and light yellow. Distances are given in angstrom and shown in red, angles are given in degree [°] and presented in black. Val-47 of both structures are shown as stick presentation. Val-47 undergoes a backbone flip from −64.5° to 154.4° when MOR04357 is bound to GM-CSF compared to the uncomplexed MOR04357. This backbone flip results in a shift of 8.2 Å of this loop toward GM-CSF along with a rotation by 39°.](/cms/asset/e6e74296-597a-4b1d-962d-04d0225316e2/kmab_a_1099774_f0003_c.gif)
Table 1. Overview table listing contact sites of MOR04357 to GM-CSF.
Figure 4. Alignment (http://www.jalview.org Citation49) of human, rhesus and rat GM-CSF amino acid sequences. Contacted residues of MOR04357 Fab on human GM-CSF are highlighted in black, supplemented by secondary structure information. Sequence identity to rhesus and rat GM-CSF in the respective regions, is depicted in gray.

Figure 5. Consensus sequences are depicted by using the Sequence Logo tool, weighing for frequency of amino acids (http://genome.tugraz.at/Logo/ Citation50). The residue numbering shown is according to Kabat. Citation27(a) CDR-H2 consensus representing 38 unique sequences. (b) CDR-L3 consensus representing 70 unique sequences.
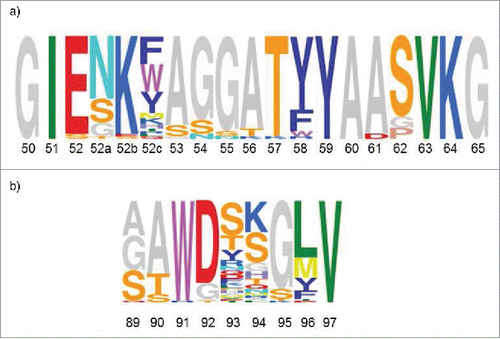
Figure 6. Stick and cartoon presentation of CDR-H2 of MOR03929 (pale blue) and MOR04252 (green) as well as GM-CSF (orange) from MOR04252:GM-CSF structure only. GM-CSF T10 from MOR03929:GM-CSF structure is shown as lines. Oxygen atoms are depicted in red, nitrogen atoms in blue. Distances are given in angstrom and depicted as dashed lines. The antibody residue numbering is according to Kabat. Citation27
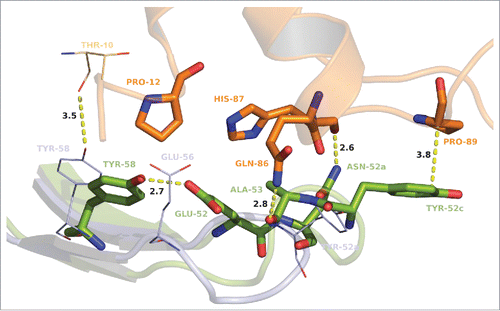
Figure 7. Stick and cartoon presentation of CDR-L3 of MOR03929 (pale blue) and MOR04302 (green) as well as GM-CSF (orange) from MOR04302:GM-CSF structure only. Oxygen atoms are depicted in red, nitrogen atoms in blue. Distances are given in angstrom and depicted as dashed lines. The antibody residue numbering is according to Kabat. Citation27
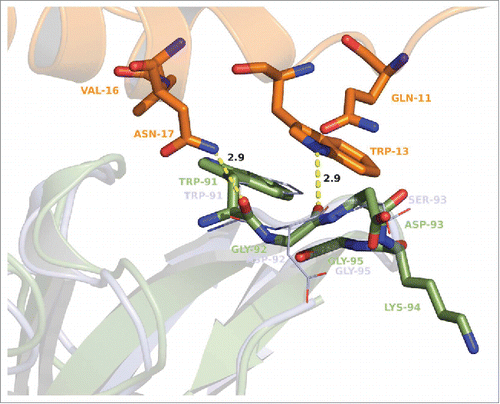
Figure 8. Superposition of GM-CSF:MOR04357 complex and GM-CSFR α/β:GM-CSF complex structure Citation16 on the basis of GM-CSF atom positions. GM-CSFR β chain is depicted in cartoon presentation as physiological dimer colored in green and violet. Incomplete GM-CSFR α chain is depicted in cartoon presentation colored in red. MOR04357 Fab molecule is presented in pale blue (Ig light chain) and dark blue (Ig heavy chain) shown as semi-transparent molecular surface. GM-CSF of MOR04357:GM-CSF complex structure is shown as cartoon presentation in orange.
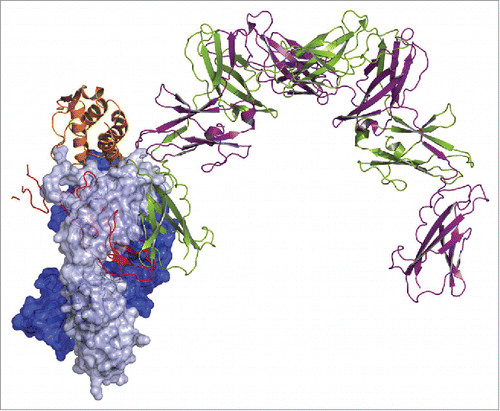
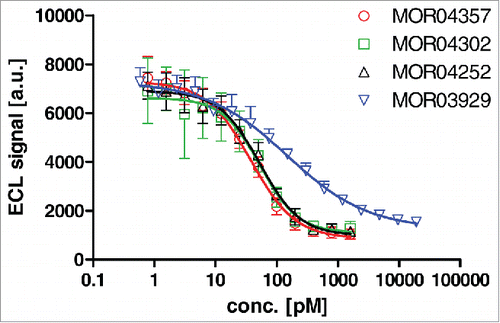
Figure 9. Dose dependent inhibition of GM-CSF binding to recombinant GM-CSF receptor α-Fc fusion protein by Fab antibodies as determined by ELISA.