Figures & data
Figure 1. Generation of mouse anti-SEMA4D (mAb 67-2) and humanized anti-SEMA4D (VX15/2503). SEMA4D KO mice were immunized with SEMA4D and mAb 67-2 was selected as the lead mouse anti-SEMA4D monoclonal antibody. This antibody was humanized and optimized to generate VX15/2503. See Materials and Methods section for details.

Figure 2. Binding of VX15/2503 to native SEMA4D expressed on human peripheral blood T lymphocytes. PBMCs from normal human blood samples were utilized as targets to assess the affinity of VX15/2503 for native, cellular SEMA4D. Using flow cytometry, fluorescently labeled beads as a standard, and modified scatchard analysis, the average KD of VX15/2503 was calculated to be 0.45 nM (SEM = 0.13). The data shown are representative of the results obtained for VX15/2503 binding to CD3+ T lymphocytes from 5 different human PBMC samples.
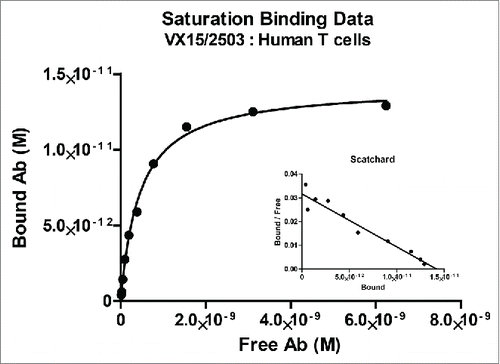
Table 1. Anti-SEMA4D Antibody Affinity for Recombinant SEMA4D1 Produced for Various Species
Figure 3. VX15/2503 functional blocking of SEMA4D with plexinB1, plexinB2, and CD72. A receptor blocking assay was developed that utilizes flow cytometry, recombinant soluble SEMA4D, and plexinB1 (A.) or plexinB2 (B.) receptor-transfected cells to quantitate blocking of SEMA4D by VX15/2503. Error bars represent intra-assay triplicates and similar assays have been repeated 2 times with similar results. (C.) An immunofluorescence blocking assay using 293.PLXNB1 and CHO.CD72 demonstrates that VX15/2503 completely blocks the binding of SEMA4D to either receptor. Transfected cells were allowed to adhere overnight, and then the antibody / recombinant his-tagged SEMA4D was added. Bound SEMA4D was detected with an anti-6xHis-APC tagged secondary antibody. Blue represents DAPI staining to visualize nuclei and red indicates SEMA4D staining. Representative images are shown and similar experiments were repeated 4x for each cell line. (D.) “Collapse” assay was performed to demonstrate the ability of VX15/2503 to block SEMA4D-induced actin rearrangement. Cells were allowed to adhere overnight and the antibody / recombinant his-tagged SEMA4D was added and then cells were fixed. Actin cytoskeleton and nuclei were stained using phalloidin-Alexa488 (green) and DAPI (blue). Representative images are shown; similar experiments have been repeated 3x with similar results.
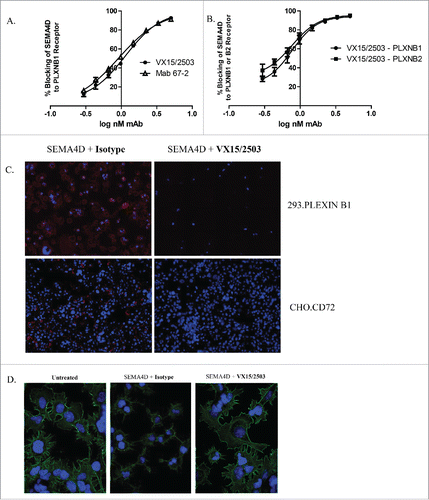
Figure 4. Characterization of VX15/2503 stability and in vitro toxicity. To assess in vitro killing, VX15/2503 was tested in non-radioactive CDC (A) and ADCC (B) assays by using alamar blue or the a-Cella tox kit, respectively. Rituximab (Rituxan®) was utilized as a positive control since the Daudi target cells were both CD20 and SEMA4D positive. To assess in vivo half antibody formation, VX15/2503 (Anti-SEMA4D S228P) or VX15/2503 without the S228P mutation (Anti-SEMA4D WT) were injected into SCID mice with additional control IgG4 antibodies (anti-C35) and serum was tested in a bispecific antibody ELISA (C) 48 hours later. (D) Human PBMCs were incubated with VX15/2503 or control mAbs and cytokines were measured using multiplex CBA analysis.
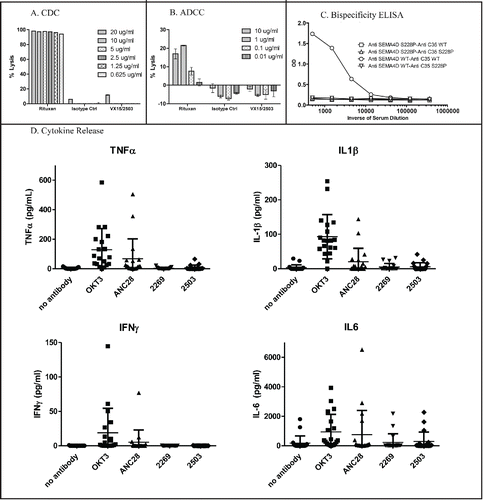
Figure 5. Epitope Mapping of VX15/2503 : SEMA4D interaction. Epitope mapping studies were performed utilizing CLIPS Epitope Mapping technology at Pepscan. Using the entire protein sequence of human SEMA4D, surface immobilized, spatially constrained peptide microarrays, and an ELISA based detection method using CDD camera quantification, the SEMA4D protein was examined for either linear or conformational epitopes of VX15/2503. Three amino-acid sequences were identified (A.) that putatively comprise the discontinuous epitope of VX15/2503. (B.) A combination ribbon / space-filling structural diagram of a SEMA4D dimer that shows the VX15/2503 epitope overlapping the dimerization and plexin-binding domains; colors indicate PLXNB1 binding site, SEMA4D dimerization domain, VX15/2503 epitope, Overlap between VX15/2503 epitope and SEMA4D dimerization domain.
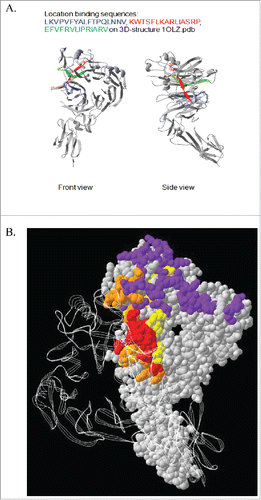
Figure 6. Internalization of VX15/2503 : SEMA4D complex upon binding cellular SEMA4D. PBMCs were incubated with Alexa 488 fluorochrome-labeled VX15/2503 or human IgG4 isotype control antibody at 37°C for up to 24 hours. The cells were washed to remove labeled antibody and incubated at 4°C in the presence or absence of an anti-Alexa 488 quenching antibody. Samples were subsequently stained with anti-human CD3-Alexa 647 to label CD3+ T cells and analyzed via flow cytometry. The quenching antibody blocks any fluorescent signal from the cell surface, so any signal remaining after quenching represents internalized antigen. A comparison was made between the GMFI values from the quenched and unquenched samples; this allowed for a calculation of the percent SEMA4D internalized using GMFI values from the Alexa 488 channel. In (A.), the % internalization data represent the mean internalization from 3 different human donors. Microscopy and imaging was performed to visualize internalization (B.); images were pseudocolored to represent DAPI (blue), CD3 (red), and VX15/2503 (green) and 2 of 4, 60X representative images are shown stained with either isotype or VX15/2503. The supernatant from the same experiment was analyzed for levels of total soluble SEMA4D (free or bound by VX15/2503) and those levels are shown for each condition in the table in part (C). The ELISA data represents the mean from 2 different human donors and similar results were obtained in a second experiment (data not shown), <LLQ indicates [sSEMA4D] <0.08 ng/ml.
![Figure 6. Internalization of VX15/2503 : SEMA4D complex upon binding cellular SEMA4D. PBMCs were incubated with Alexa 488 fluorochrome-labeled VX15/2503 or human IgG4 isotype control antibody at 37°C for up to 24 hours. The cells were washed to remove labeled antibody and incubated at 4°C in the presence or absence of an anti-Alexa 488 quenching antibody. Samples were subsequently stained with anti-human CD3-Alexa 647 to label CD3+ T cells and analyzed via flow cytometry. The quenching antibody blocks any fluorescent signal from the cell surface, so any signal remaining after quenching represents internalized antigen. A comparison was made between the GMFI values from the quenched and unquenched samples; this allowed for a calculation of the percent SEMA4D internalized using GMFI values from the Alexa 488 channel. In (A.), the % internalization data represent the mean internalization from 3 different human donors. Microscopy and imaging was performed to visualize internalization (B.); images were pseudocolored to represent DAPI (blue), CD3 (red), and VX15/2503 (green) and 2 of 4, 60X representative images are shown stained with either isotype or VX15/2503. The supernatant from the same experiment was analyzed for levels of total soluble SEMA4D (free or bound by VX15/2503) and those levels are shown for each condition in the table in part (C). The ELISA data represents the mean from 2 different human donors and similar results were obtained in a second experiment (data not shown), <LLQ indicates [sSEMA4D] <0.08 ng/ml.](/cms/asset/00bba257-f704-4569-b497-7051c5660ef8/kmab_a_1102813_f0006_c.gif)
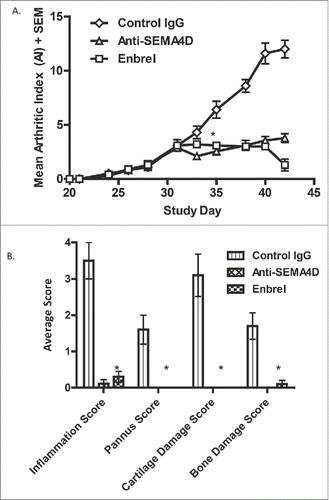
Figure 7. Treatment with anti-SEMA4D inhibits progression in a murine model of collagen-induced arthritis. (A.)To induce disease, DBA/1J mice were immunized subcutaneously (s.c.) on day 0 with an emulsion of collagen / CFA and challenged (s.c.) on day 21 with collagen / IFA. Treatment with either control IgG, mAb 67-2 (anti-SEMA4D), or Enbrel® 2x/week was started after mice reached an AI of 3 (day 31). (B.) Mice were sacrificed at endpoint (day 42) and paws were fixed, stained with toluidine blue and analyzed and scored histologically as described in the Material and Methods section. * Error bars represent S.E.M. and significance was calculated versus the control Ig group using Student's t-test, p≤0.001 for both Anti-SEMA4D and Enbrel groups for day 35 and later in 7A and at day 42 in 7B.