Figures & data
Table 1. Soluble Panning Results. Number of binders identified from soluble panning using biotinylated TGFβ2. Comparative data is provided for both XOMA phage display library formats: the human Fab library (XFab1) and the human single chain antibody, scFv, (XscFv2).
Table 2. Sequence Diversity of Binders. Number of unique sequences from binders to more than one TGFβ isoform.
Figure 1. Sequence Comparison of Parent XPA.42.068, Affinity Matured Variant XPA.42.681, and XPA.42.089. The heavy and light chain sequence of the parent XPA.42.068 is compared to the affinity improved light chain variant, XPA.42.681. Amino acid positions that are different between these clones are highlighted in bold black type. For XPA.42.089, amino acids are highlighted which are different than the XPA.42.068 clone.
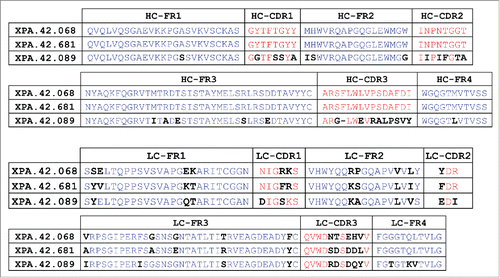
Figure 2. Antibody Neutralization of TGFβ Mediated HT-2 Growth Arrest. HT-2 cells were treated with a fixed dose of TGFβ1, 2, or 3 (EC80 level) in the presence of neutralizing antibody titrations. Cell growth was measured after 48 hours of incubation at 37°C using a luminescent cell-viability reagent (CellTiter-Glo®). Values are presented as percent of a no TGFβ control.
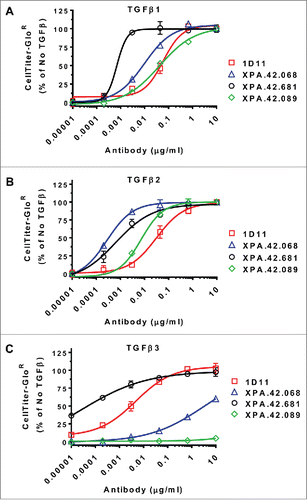
Figure 3. Antibody Neutralization of TGFβ Stimulated IL-11 release from A549 Cells. A549 cells were treated with a fixed dose of TGFβ1, 2, or 3 (EC80 level) in the presence of neutralizing antibody titrations. After 24 hours at 37°C, IL-11 levels in cell culture supernatants were measured by ELISA. Values are reported as percent of a no antibody control.
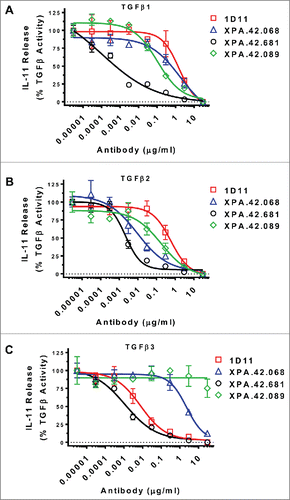
Table 3. Antibody Affinity Using IgG Capture Method. Antibodies were immobilized on a CM5 sensor chip of a Biacore 2000 via amine chemistry. TGFβ was injected at 10 nM, 2 nM, 0.4 nM, and 0.08 nM. Data were analyzed for binding rate parameters using the Scrubber software after double referencing.
Table 4. Antibody Affinity Using Immobilized TGFβ. TGFβs were immobilized on a CM5 chip via amine chemistry. Antibodies were then injected at 33.33 nM, 6.67 nM, 1.33 nM, 267 pM, and 53 pM. Data were analyzed for binding rate parameters using the Scrubber software after double referencing.
Figure 4. Antibody Affinity Using Immobilized IgG Method. Antibodies were immobilized on a CM5 sensor chip of a Biacore 2000 via amine chemistry. TGFβ was injected at 10 nM, 2 nM, 0.4 nM, and 0.08 nM. Data were analyzed for binding rate parameters using the Scrubber software after double referencing.
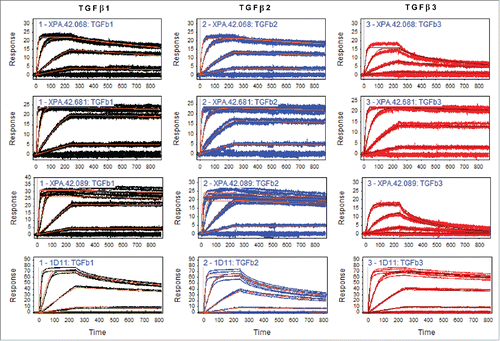
Figure 5. TβRII and TβRIII competition by antibodies utilizing TGFβ1, TGFβ2, and TGFβ3. TGFβRII or TGFβRIII protein was immobilized on a SPR surface. Antibodies were incubated at various concentrations with 4 nM TGFβ2 or 1.6 nM TGFβ3 prior to injection over the receptor-coated surfaces. Binding levels at the end of a 2 minute injection were recorded and values were normalized to a no antibody control samples. The data were fit using a 4 parameter fit in GraphPad Prism.
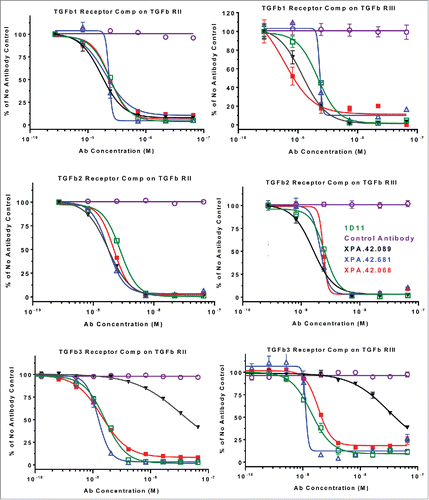
Figure 6. Antibody competition with TGFβ1 binding to rhLAP. rhLAP was immobilized on a CM5 sensor chip of a Biacore 2000. TGFβ1 was injected at 10 nM in the presence of 66.6 nM IgG. Level of TGFβ1 bound to rhLAP at the end of the injection is shown. A lack of signal shows competition with TGFβ1 binding by the antibodies.
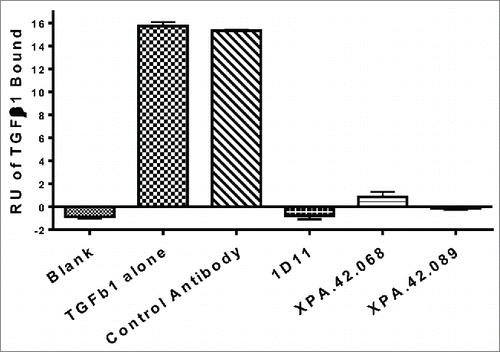
Table 5. Stoichiometry evaluation: Data summary from antibody capture kinetics RUmax test using TGFβ2. Antibodies were captured at 107-117 RU onto an anti-human-Fc capture surface and TGFβ2 was injected in duplicate at 20 nM, 4 nM, and 0.8nM. Data was then double referenced and fit kinetically to establish the RUmax using the formula TGFβ/antibody = [TGFβ RU/(Ab RU/6)].
Figure 7. Inhibition of SMAD2 phosphorylation. Detroit 562 cells were pre-incubated with neutralizing or control antibodies (50 ng/mL), then stimulated with 5 ng/ML TGFβ1, -2, or -3 for 30 minutes. Cell lysate were prepared and analyzed by ELISA for total and phosphorylated SMAD2. Percent inhibition of pSMAD2 was normalized to total SMAD2 for each clone relative to the anti-KLH control antibody.
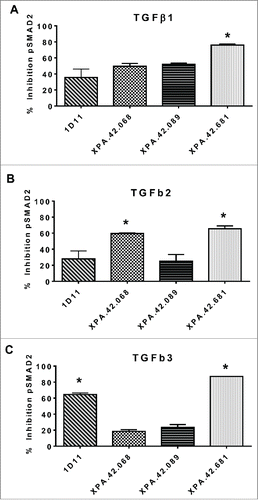
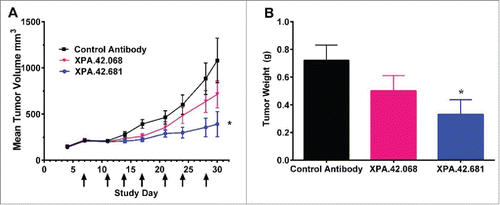
Figure 8. Detroit 562 xenograft tumor volumes following twice weekly IP injections of XPA.42.068, XPA.42.681 or control antibody.Animals were implanted at Day 0 and randomized at Day 4 following establishment of tumors. Each group with 12 mice per group was treated twice weekly with 3mg/kg XPA.42.068, XPA.42.681 or isotype control antibody. Tumor volumes were determined prior to dosing. A) Change in mean tumor volume over time for each dose group. Arrows indicate days of antibody injection. Control Antibody (squares), XPA.42.068 (inverted triangles), and XPA.42.681 (Circles) B). Mean tumor weight for each group taken at Day 30 post-implantation. (*=p < 0.05).