Figures & data
Figure 1. Affinity comparison of mAbs derived from chicken immunizations and other sources for 2 unrelated model antigens, PCSK9 and PGRN. (A) Biacore binding curves and global fits of select anti-PCSK9 mAbs from chicken immunization showing a diverse set of kinetic profiles. The colored curves represent the measured binding responses of hPCSK9 when injected at concentrations of 5 nM (red) and 50 nM (green), with the global fit overlaid in black. (B) Isoaffinity plot comparing anti-PCSK9 mAbs generated from chicken immunization (olive green) with those from human phage display libraries (blue). (C) Isoaffinity plot comparing anti-PGRN mAbs generated from immunizations in chicken (olive green) and mouse (purple). The red dotted line indicates the kd limit of 5.70 × 10−5 (1/s) that was placed on interactions which showed < 5 % signal decay within the allowed dissociation phase of 15 min, also known as the “5 % rule” (see Methods).Citation20
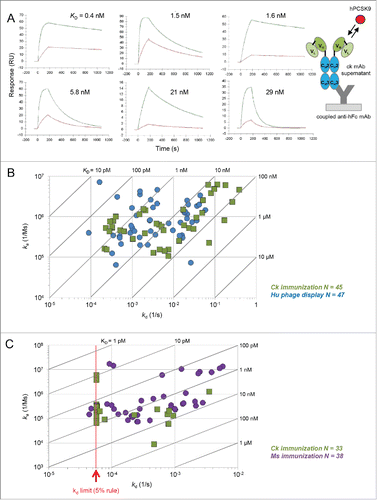
Figure 2. Solution affinity determination of 2 high affinity clones obtained from chicken immunization toward their respective serum antigens. (A) Schematic representation of the KinExA assay set-up. The mAb of interest is titrated into human serum and the equilibrated mixtures are injected over beads absorption-coated with a competing mAb to capture the free antigen. The bead-captured antigen is then detected using a Dylight-labeled sandwiching mAb. (B) Global analysis of mAb C34 binding serum PCSK9. (C) Global analysis of mAb C25 binding serum PGRN. In each case, the reported apparent KD value is the best fit and 95 % confidence interval of the fit.
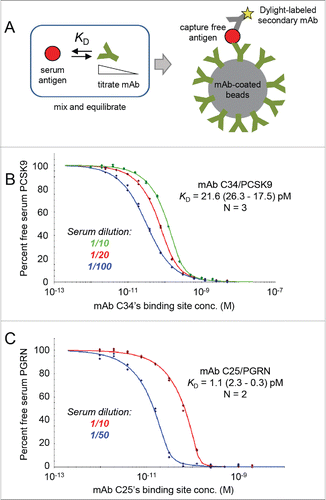
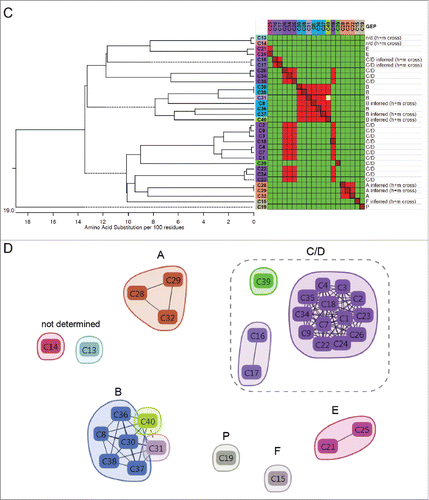
Figure 3. Chicken-human merged binning results for PCSK9. (A) Heat map showing binning assignments for 39 mAbs generated from chicken immunization (olive green) and 24 mAbs generated by human phage display (blue). SPR-derived KD values toward human PCSK9 are reported (conditionally formatted using a color gradient), along with their Octet-based cross-reaction toward human (h), mouse (m) and rat (r) PCSK9 (n/d = not determined). (B) Dendrogram showing antibody sequence lineages of the chicken mAbs alongside the binning heat map for these clones (drawn from panel A, transposed, and resorted). See Table S1.
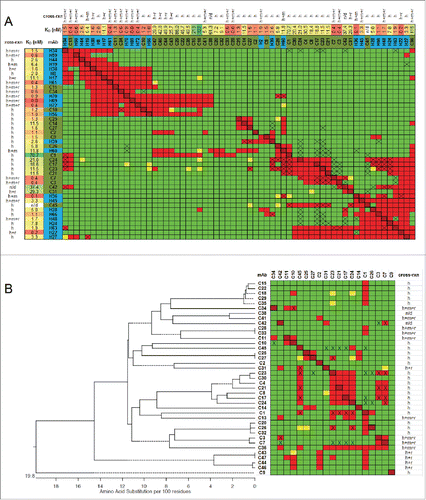
Figure 4. Chicken-mouse merged binning results for PGRN. (A) Heat map showing binning assignments for 32 mAbs generated from chicken immunization (olive green) and 20 mAbs generated from mouse immunization (purple). SPR-derived KD values toward human PGRN are reported, along with their cross-reaction toward mouse (m) PGRN, and their GEP subdomain assignment (just for the human-specific clones; n/d = not determined). (B) Blocking network plot, colored by mAb library, with GEP assignments indicated (determined empirically, or inferred). (C) Dendrogram of the antibody sequence lineages for the chicken mAbs alongside the binning heat map for these clones (drawn from panel A, transposed, and resorted). (D) Blocking network plot for the chicken clones, colored by bin, with GEP assignments indicated. See Table S2.
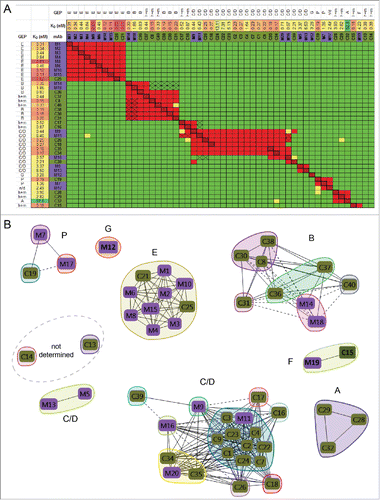
Table 1. Comparison of the epitope coverage observed for anti-PGRN mAbs raised via the immunization of chickens or mouse. *Of the 14 chicken mAbs that showed human/mouse crossreactivity, 8 were mapped to a GEP subdomain (by inferring from the binning data shown in ) and are included in the A – P tally, and 6 were not assigned to a GEP subdomain.