Figures & data
Figure 1. Schematic representation of the workflow used for determination of the light and heavy chain variable domain sequences.
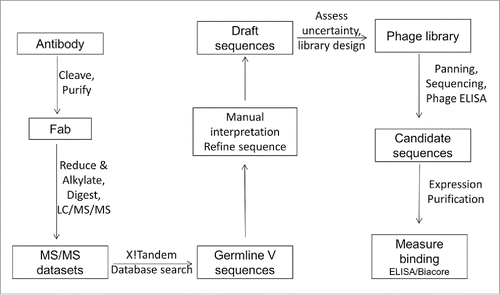
Figure 2. Mass spectra and location of protease cleavage sites during LOB12.3 and 3H3 Fab generation. (A) Intact mass spectra of reduced Fab generated by papain digestion of LOB12.3 IgG. The rat IgG1 hinge region sequence is shown above the spectra and sites of cleavage by papain (open triangles) or SpeB (filled triangle) are shown. (B) Intact mass spectra of reduced Fab generated by papain digestion of 3H3 IgG. The rat IgG2a hinge region sequence is shown above the spectra and sites of cleavage by papain are shown.
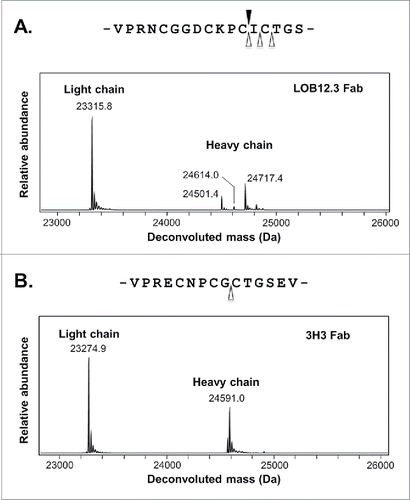
Figure 3. LOB12.3 variable domain sequences described in this work. LOB12.3-Lv1/Hv1 represent the initial assembly of proteomic data into light and heavy V-domain sequences, where X represents ambiguous regions of sequence (gray highlighting = 113 Da; green = 114 Da). Alignment of this draft to closest V, D and J translated germline gene segments is shown below. LOB12.3-Lv2/Hv2 were refined draft sequences, as defined in , and ambiguous regions in this light/heavy pair were programmed into a Fab phage display library. Further refinement after phage display led to LOB12.3-Lv3/Hv3, and finally 2 unique light/heavy pairings (LOB12.3-Lv4/Hv4 and LOB12.3-Lv4/Hv5) that were part of recombinant Fab fragments. Sequence numbering is according to Kabat et al.Citation18 CDRs are shown in bold and were defined according to the sequence definition of Kabat et al.,Citation18 except for CDR-H1, which is the combined sequence and structural definition.Citation19

Table 1. Ambiguous regions of sequence and strategy for resolving these.
Figure 4. MS/MS spectrum of a 3H3 chymotryptic peptide containing part of CDR-H3. Interpretation of the fragmentation observed from the 728.3 Da parent ion led to assignment of the sequence as CT(L/I)DGY where C is modified with a carbamidomethyl group. Additional peaks arise primarily by water loss from the labeled b ion peaks.
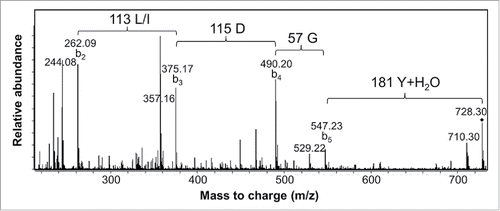
Figure 5. 3H3 variable domain sequences described in this work. 3H3-Lv1/Hv1 represent the initial assembly of proteomic data into light and heavy V-domain sequences, where X represents ambiguous regions of sequence (gray highlighting = 113 Da; yellow = 158 Da; blue = 216 Da; green = 114 Da; magenta = 144 Da). An alignment of this draft to closest V and J translated germline gene segments is show below (a germline D-segment could not be assigned based on the initial draft sequence). 3H3-Lv2/Hv2 were refined draft sequences, as described in , and ambiguous regions in this light/heavy pair were programmed into 2 Fab phage display libraries. Further refinement after phage display led to 3H3-Lv2/Hv3, and finally 2 unique light/heavy pairings (3H3-Lv2/Hv4 and 3H3-Lv2/Hv5) that were part of recombinant Fabs. Sequence numbering is according to Kabat et al.Citation18 CDRs are shown in bold and were defined according to the sequence definition of Kabat et al.,Citation18 except for CDR-H1, which is the combined sequence and structural definition.Citation19

Table 2. Distribution of sequences following 2 (R2) or 3 (R3) rounds of phage display selection.
Figure 6. Binding of reverse-engineered LOB12.3 and 3H3 Fab variants to murine CD137. A) Competition phage ELISA measuring relative CD137 binding affinities to phage displayed LOB12.3 or 3H3 Fab variants. LOB12.3 Fab variants contained LOB12.3-Lv3/Hv3 light/heavy V-region pairs, where the sequence at VL33/VH100c was Leu/Leu (green), Leu/Ile (blue), Ile/Leu (magenta) or Ile/Ile (red). IC50 values for displacement of 50% of bound Fab-phage was between 2.0 – 3.9 nM for all 4 variants. 3H3 Fab variants contained 3H3-Lv2/Hv3 V-region pairs, where the sequence at VH29 was Ile (blue), or contained a VH-E61D substitution and Ile (red) or Leu (magenta) at VH29. IC50 values for displacement of 50% of bound Fab-phage was between 3.1 – 6.2 nM for all 3 variants. B) Sensorgram traces showing near identical binding kinetics for 11.1 nM injections of Fab samples over an immobilized CD137 surface. Reverse-engineered Fab variants (blue = Fab1; green = Fab2) are compared to the corresponding reference Fabs generated by partial proteolysis of the commercially-sourced parental IgG (black = Fabp). Binding kinetics calculated from the full set of surface plasmon resonance data are shown in .
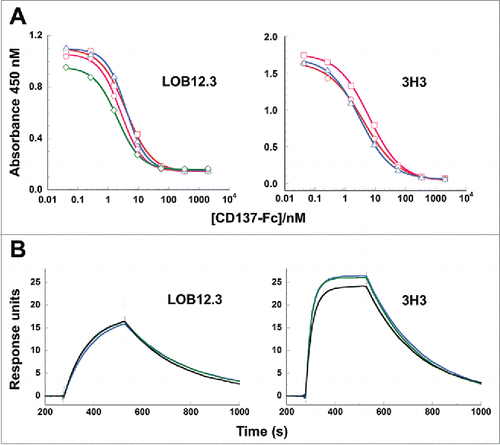
Table 3. Binding affinities of reverse-engineered 3H3 and LOB12.3 Fab variants to CD137.
