Figures & data
Figure 1. Schematic view of lectin microarray. (A) Lectin microchips used in this study consist of 45 distinct lectins that selectively bind structural variants of carbohydrates attached onto a protein. Each lectin is printed in triplicate. The lectin-printing layout of lectin chips was provided by the vendor (GlycoTechnica). (B) Protein samples are labeled with a fluorescent dye (e.g., Cy3) and then applied onto the lectin chips. The binding signals at each lectin spots are measured using an evanescent-field fluorescence scanner, detecting the presence or absence of glycan variants in the testing sample based on the known selectivity of lectins toward particular glycan structures.
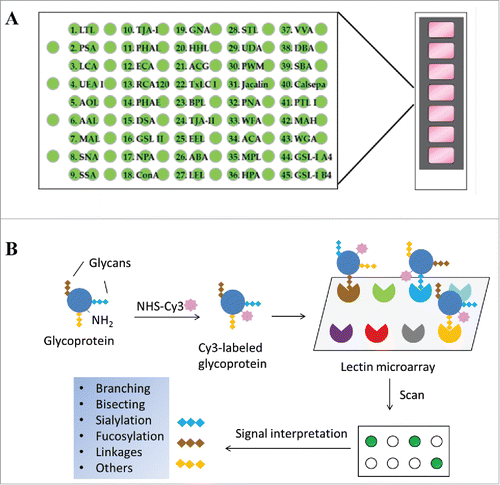
Table 1. Reported glycan selectivity of the 45 lectins used in the microarray assay.*Citation16
Table 2. Information of protein samples used in lectin microarray assay.*
Figure 2. Lectin binding profiles of therapeutic IgG monoclonal antibodies. The indicated therapeutic mAbs, including bevacizumab, trastuzumab, adalimumab, infliximab, rituximab, omalizumab, and panitumumab were labeled with Cy3 and applied onto the lectin chips containing 45 distinct lectins with each being printed in triplicate. (A) Lectin binding images of the indicated samples. (B) Relative binding signals at specific lectin spots were derived from the images in A and normalized to protein markers on the same chip (mean ± SD). Shown are representatives of 3 independent experiments. The coefficient of variation (CV) was determined to be < 10% for most lectin-glycan binding signals of the samples tested. (C) Typical glycan structures present in the Fc portion of therapeutic IgG1 mAbs.Citation27
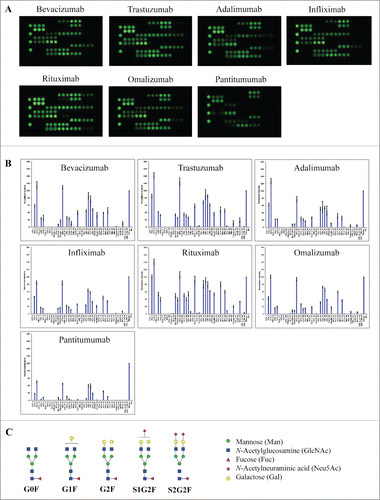
Figure 3. Lectin binding profiles of cetuximab and etanercept. Cy3-labeled samples were applied onto the lectin chips as in . Shown are (A) representative lectin binding images, (B) Relative binding signals at specific lectin spots (mean ± SD), and (C) Typical glycan structures present in the Fab portion of cetuximab.Citation30
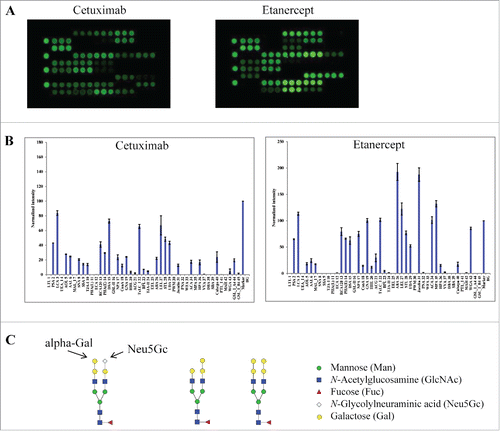
Figure 4. Glycan profiling of Fabs and Fcs. The Fabs and Fcs of rituximab (A) and cetuximab (B) were prepared as described in Materials and Methods. Purified Fab and Fc were analyzed by reducing SDS-PAGE (left panel) and lectin microarray (right panel). As noted, the dimeric Fcs (˜ 55 kDa) were reduced to monomeric products (˜ 30 kDa) on SDS-PAGE under reducing conditions.
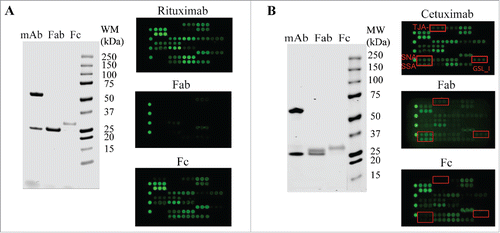
Figure 5. Glycan profiles of proteins produced by different host cell systems. The proteins tested include therapeutic proteins produced by CHO cells (darbepoetin alfa and dornase alfa), yeast strains (rasburicase), or E. coli (filgrastim), and human transferrin protein expressed by recombinant rice (transferrin-rice) or isolated from human plasma (transferrin-human). (A) Lectin binding images. (B) Relative binding signals at specific lectin spots (mean ± SD). (C) Typical glycan structures present in darbepoetin alfa.Citation36
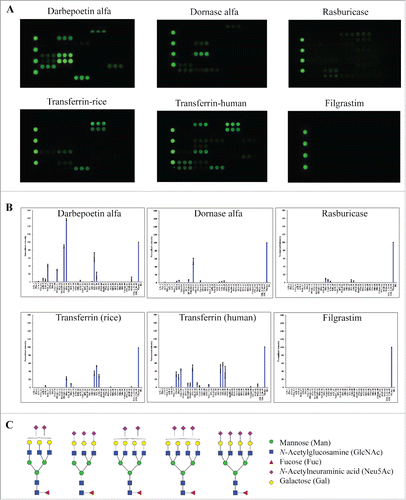
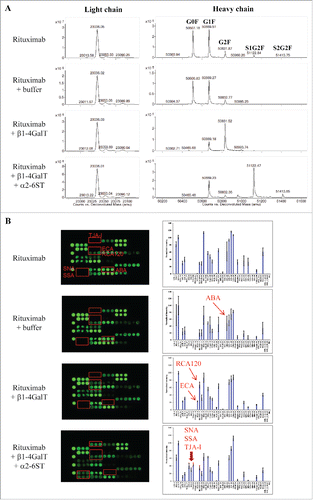
Figure 6. Assessing glycan variants in glycan-engineered rituximab protein samples. Rituximab was incubated with a reaction buffer alone (rituximab + buffer), β1-4-galactosyltransferase (rituximab + β1-4GalT), and β1-4-galactosyltransferase followed by α2-6-sialyltransferase (rituximab + β1-4GalT + α2-6SiaT) (see detail in Materials and Methods). After affinity purification, the resulting samples were analyzed using mass spectrometry and lectin microarray, respectively. Shown are representatives of deconvoluted mass spectra (A) and corresponding lectin binding profiles (B) for the samples produced under the indicated conditions.