Figures & data
Figure 1. Calculation of KD by interpolation of equilibrium fit. Sensorgrams (a) and derived affinity plots (b) of human IgG1 anti-RhD binding to ligands FcγRI, FcγRIIb, FcγRIII and FcγRIV for 4 different ligand (FcγR) concentrations; 30, 10, 3 and 1nM, c) The affinity plot derived KD and Rmax of each ligand concentration are plotted for interpolation of KD to a constant Rmax of 500.
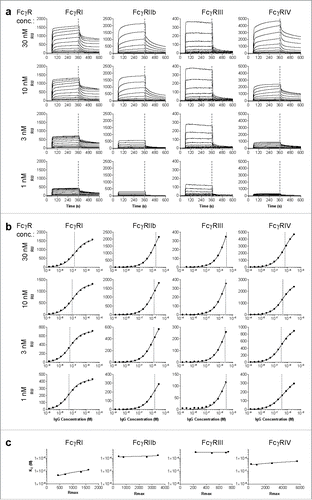
Figure 2. Binding of mouse IgG subclasses to mouse FcγR. The mouse IgG1, IgG2a, IgG2b and IgG3 with anti-Kell specificity were assessed for their binding affinity in KD (M) to the mouse FcγRI, FcγRIIb, FcγRIII and FcγRIV by SPR. −/− denotes no binding detected. n = 3. The horizontal dashed line represents the maximum concentration of IgG which is used, and the threshold required to accurately calculate the KD.
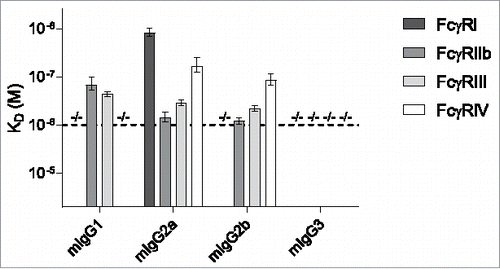
Figure 3. Binding of human IgG subclasses to mouse FcγR. The human IgG1, IgG2, IgG3 and IgG4 with anti-RhD specificity (clone 19A10) were assessed for their binding affinity in KD (M) to the mouse FcγRI, FcγRIIb, FcγRIII and FcγRIV by SPR. −/− means no binding detected; *denotes binding > 2 μM or > 3 μM for hIgG2 or hIgG4, respectively, represented by the horizontal dashed lines, which represent the maximum concentration of IgG that is used, and the threshold required to accurately calculate the KD, 2 μM for IgG2 and 3 μM for IgG1, IgG3 and IgG4; **KDs for FcγRIII binding of hIgG1, hIgG2 and hIgG4 represent the average of 2 alternative estimates (see Material & Methods section for details). n = 3.
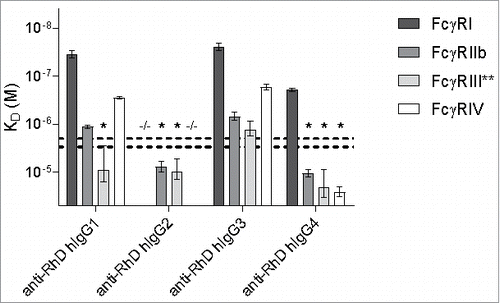
Table 1. Affinity in KD (μM) of human IgG subclasses for mouse FcγRs, SEM in brackets behind KD, −/− no binding detected.
