Figures & data
Figure 1. CALY-002 epitope and mode of action. (A) CALY-002 does not prevent IL-15 binding to IL-15Rα. Biotinylated hIL-15 at a single concentration was incubated with immobilized IL-15Rα-Fc in the presence of a titration of antibodies, as described. Data represent mean ± SD of IL-15 binding signal expressed as 450 nM absorbance from one representative experiment. (B) The published structure of the IL-15/IL-15Rαβγ quaternary complex was used to visualize the IL-15 binding epitope of CALY-002. Interleukin-15 is shown in blue, the IL-15 binding motif of CALY-002 in cyan, and IL-15Rβ in orange. The side chain of the critical residues of the binding motif of CALY-002 is highlighted: D109, E112, I116, and N119. K71 of IL-15Rβ directly interacts with D109 of IL-15.
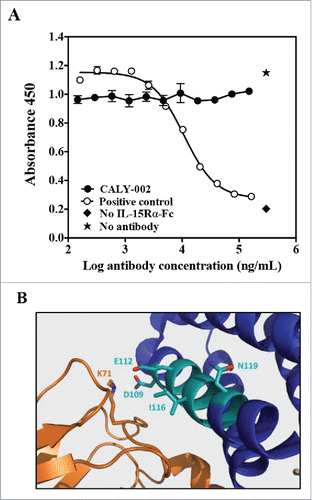
Table 1. Characterization of the binding affinity, species specificity and selectivity of CALY-002. Kinetic interaction of CALY-002 with recombinant IL-15 was measured by surface plasmon resonance. Dissociation (koff) and association (kon) rate constants were obtained by nonlinear regression analysis. KD = koff /kon. Selectivity and selectivity of CALY-002 toward recombinant human, monkey, mouse and rat IL-15 as well as human IL-2 was tested by ELISA. Whenever appropriate, the half-maximum binding concentration of CALY-002 (BC50) was calculated. Results are expressed as the mean of 3 independent experiments ± SD.
Table 2. CALY-002 inhibits IL-15-mediated cell proliferation. The effect of CALY-002 on the proliferation and survival of M-07e and Kit 225 cells induced by recombinant soluble human or monkey IL-15, or human IL-15 immobilized onto recombinant human IL-15Rα-Fc, was measured as described in Materials and Methods. The half-maximum inhibitory concentration (IC50) of CALY-002 in each assay was determined and results are expressed as mean IC50 ± SD for at least 3 experiments for each condition tested.
Figure 2. Pharmacodynamic inhibition of IL-15 by in vivo in mice and monkeys. (A) NK cell expansion was induced in groups of 5 mice by injections of hIL-15 mixed with hIL-15Rα-Fc at Day 1, 2 and 3. PBS (gray bar), CALY-002 (black bar) or control isotype antibody (dashed bar) were injected 20 minutes before the first injection of hIL-15/hIL-15Rα-Fc. Animals were killed at Day 4 and number of NK cells present in individual spleens were determined. Naive mice were injected with saline alone (no hIL-15/hIL-15Rα-Fc, white bar). Data are plotted as mean ± SD. Statistical analysis: Student's t test. (B) Serum levels of human IgG in cynomolgus monkeys receiving a single i.v. dose of 10 mg/kg (closed triangles), 1 mg/kg (open circles) or 0.1 mg/kg (closed circles) CALY-002. Results are expressed as mean ± SD for 3 to 4 animals per group. (C) CALY-002 reduces circulating NK cell numbers in cynomolgus monkeys. Following a single dose injection of 10 mg/kg (closed triangles), 1 mg/kg (open circles) or 0.1 mg/kg (closed circles) CALY-002, animals underwent a 4-wk observation period and NK cell counts were determined by flow cytometry at Day −3 (pre-dose), and 1, 3, 5, 8, 14, 21 and 28 d after treatment. Data are plotted as mean absolute number of blood CD3−CD16+ cells NK cells ± SD for 3 to 4 animals per group.
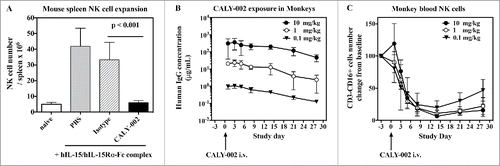
Figure 3. CALY-002 inhibits IL-15–induced survival and STAT5 phosphorylation of type II RCD patient IEL cell lines. Type II RCD IEL cell lines were cultured with medium (white bars), IL-15 (black bars), IL-15 with CALY-002 (hatched bars) or IL-15 with control isotype (gray bars) for 48h. (A) Percentages of apoptotic cells in cultured IELs were determined by labeling using an AnnexinV-APC/PtdIns Kit and (B) phosphorylated STAT5 was stained with PEcy7-labeled anti-p-STAT5 antibody. Results were transformed as percentage of control (medium) for the percentage of apoptotic cells or the MFI of phosphorylated STAT5 intracellular expression induced by IL-15, and expressed as mean ± SD of results obtained with 3 different type II RCD IEL cell lines.
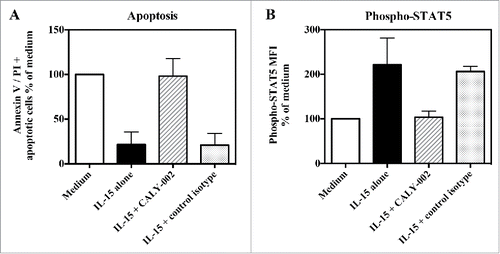
Figure 4. CALY-002 effect on spleen and intestinal intra-epithelial lymphocyte subsets in IL-15 transgenic mice. Total number of (A) spleen and intestinal intra-epithelial (B) CD3+CD8+, CD3+CD4+, CD3−CD19+, CD45+CD3−NK1.1 and TCRαβ+NK1.1+ lymphocytes at Day 28. Symbols represent individual IL-15 transgenic mice treated with either CALY-002 (closed circles) or control isotype (open squares) antibodies, or control C57BL/6 mice (closed triangles). Results are expressed as mean ± SD of absolute cell number/μL for a 1 mL suspension per animal. Statistical analysis: Student's t test. *p < 0.05; **p < 0.01; ***p < 0.001; ns: not significant.
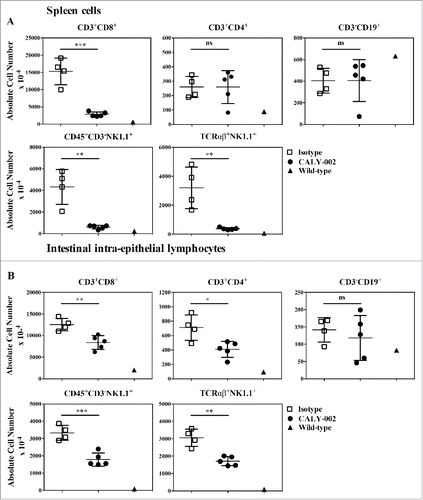
Figure 5. Esophageal biopsies from active EoE patients show increased IL-15 and IL-15Rα gene expression, and IL-15 expression correlates with EoE molecular score. RNA was extracted from esophageal biopsies, and subjected to quantitative RT-PCR gene expression analysis. (A) IL-15 mRNA expression, (B) IL-15Rα mRNA expression and (C) EoE molecular score from control healthy subjects (closed circles), active EoE non-treated patients (open circles), inactive EoE corticoid-responder patients (closed diamonds), and active EoE corticoid non-responder patients (open diamonds). Each symbol represents a single biopsy and mean ± 95% CI is indicated for each group. Statistical analysis: Student's t test. *p < 0.05; **p < 0.01; ***p < 0.001, ****p < 0.0001. (D) Correlation between EoE molecular score and IL-15 mRNA expression for all subjects. Each symbol represents a single biopsy.
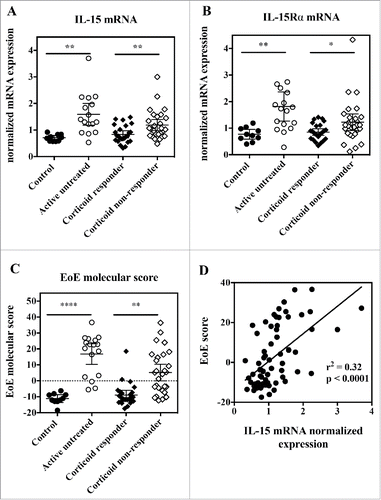
Figure 6. Increased IL-15 protein expression in esophageal biopsies from active EoE patients. (A) Immunofluorescence. Staining for IL-15 (green) and CD3 (red) was performed in 4 active EoE patients [4 x proximal and 4 x distal tissue samples], 2 normal control individuals [2 x proximal and 2 x distal tissue samples], 3 corticosteroid responders [3 x proximal and 3 x distal tissue samples] and 2 corticosteroid non-responders [2 x proximal and 2 x distal tissue samples]. Representative images are shown for each group. White arrows indicate examples of double stained cells, and filled white triangles indicate T cells without detectable IL-15 staining. Scale bars, 10 μm. Quantification of IL- 15 expression in epithelial cells is shown in the left panel. The right panel shows the quantitative analysis of infiltrating T cells that were either IL-15 negative (black bars) or positive (gray bars). (B) Immunohistochemistry. Staining for IL-15 (red) and ECP (brown) in active EoE, normal control individuals, corticosteroid responders, and corticosteroid non-responders (same esophageal tissues as described in panel A). Representative images are shown for each group. Insets demonstrate infiltrating eosinophils that are positive for IL-15 (black arrows) or IL-15 negative (filled black triangles). Scale bars, 10 μm. The right panel shows the quantitative analysis of infiltrating eosinophils that were either IL-15 negative (black bar) or positive (gray bar). Statistical analysis: one-way ANOVA followed by Tukey's multiple comparisons. ****p < 0.0001; ***p < 0.001; **p < 0.01; *p < 0.05.
![Figure 6. Increased IL-15 protein expression in esophageal biopsies from active EoE patients. (A) Immunofluorescence. Staining for IL-15 (green) and CD3 (red) was performed in 4 active EoE patients [4 x proximal and 4 x distal tissue samples], 2 normal control individuals [2 x proximal and 2 x distal tissue samples], 3 corticosteroid responders [3 x proximal and 3 x distal tissue samples] and 2 corticosteroid non-responders [2 x proximal and 2 x distal tissue samples]. Representative images are shown for each group. White arrows indicate examples of double stained cells, and filled white triangles indicate T cells without detectable IL-15 staining. Scale bars, 10 μm. Quantification of IL- 15 expression in epithelial cells is shown in the left panel. The right panel shows the quantitative analysis of infiltrating T cells that were either IL-15 negative (black bars) or positive (gray bars). (B) Immunohistochemistry. Staining for IL-15 (red) and ECP (brown) in active EoE, normal control individuals, corticosteroid responders, and corticosteroid non-responders (same esophageal tissues as described in panel A). Representative images are shown for each group. Insets demonstrate infiltrating eosinophils that are positive for IL-15 (black arrows) or IL-15 negative (filled black triangles). Scale bars, 10 μm. The right panel shows the quantitative analysis of infiltrating eosinophils that were either IL-15 negative (black bar) or positive (gray bar). Statistical analysis: one-way ANOVA followed by Tukey's multiple comparisons. ****p < 0.0001; ***p < 0.001; **p < 0.01; *p < 0.05.](/cms/asset/5f477c1e-4a9f-4d74-8719-928416555df9/kmab_a_1332553_f0006_oc.gif)
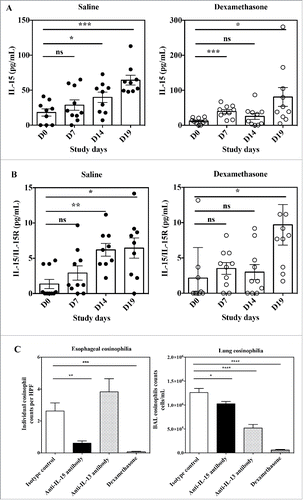
Figure 7. Effects of anti-IL-15 monoclonal antibody in the Aspergillus fumigatus mouse model of EoE. Serum concentration of free IL-15 (A) and IL-15/IL-15Rα heterodimer (B) in mice at Day 0 (before antigen challenge) and study Days 7, 14, and 19 of repeated intranasal challenge with Aspergillus fumigatus. Groups of 10 mice were treated either with saline control (closed circles) or dexamethasone (open circles). Data show individual and group mean ± SEM data for each group of animals at each time point. Statistical analysis: Student's t test. (C) Individual high power field (HPF) esophageal eosinophil counts (left panel) and total numbers of eosinophils in BAL fluid (right panel) of mice at Day 19 of repeated intranasal challenge with Aspergillus fumigatus and treated with control isotype antibody (white bar), anti-IL-15 antibody (black bar), anti-IL-13 antibody (dashed bar) or dexamethasone (hatched bar). Data are expressed as mean ± SEM for each group of animals. Statistical analysis: one-way ANOVA followed by Tukey's multiple comparisons. ****p < 0.0001; ***p < 0.001; **p < 0.01; *p < 0.05.