Figures & data
Figure 1. 1.2 L bioreactor fed-batch experiment using a feed containing either cysteine (control, n = 2), cysteine stabilized with pyruvate (CP) or SSC (n = 5 each). Feed was added at 3% (v/v) at day 3 and 6% (v/v) at days 5, 7, 9 and 14. (A) Viable cell density (plain lines) and viability (dotted lines), (B) Integral viable cell density, (C) IgG concentration in the supernatant measured by a turbidometric method, (D) Specific productivity.
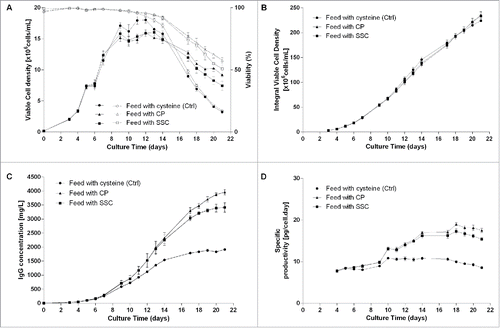
Figure 2. Overall cell specific productivity for mAb1 to 3 in cysteine, CP/CKG and SSC-containing processes. The overall specific productivity was determined by calculating the slope from the linear regression between titer and corrected integral VCD. Values are mean ± SEM obtained from 4 independent replicates. A p-value of less than 0.05 (*) was considered significant (Mann-Whitney test).
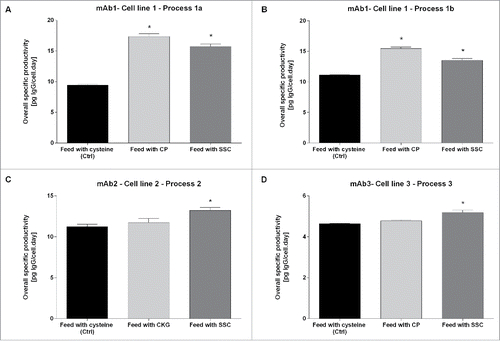
Figure 3. (A) Overlay of extracted ion chromatograms showing the relation between the LC-HC linkage with a disulfide bridge (elution at 5.18 min) and the LC-HC link with a trisulfide bridge (elution at 6.05 min). The black line corresponds to the control condition using cysteine-containing feed, light gray to the CP-containing feed and dark gray to the SSC-containing feed. (B) Mass spectra of the 3 samples showing the isotopic pattern compared with the theory and the mass accuracy. (C) Example of fragment ion spectrum of the trisulfide linked LC-HC peptide with annotation of the main peaks.
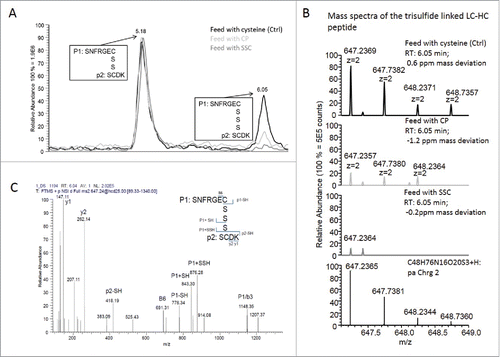
Figure 4. Relative quantification of the amount of trisulfide linkages between light chain and heavy chain of the IgG compared with the disulfide linkages quantified in 4 different monoclonal antibodies or processes. (A) Trisulfide content in mAb1 at day 13 and day 18 of the fed-batch process 1a. (B) Trisulfide content in mAb1 at day 12 of the fed-batch process 1b. (C) Trisulfide content in mAb2 at day 13. (D) Trisulfide content in mAb3 at day 14.
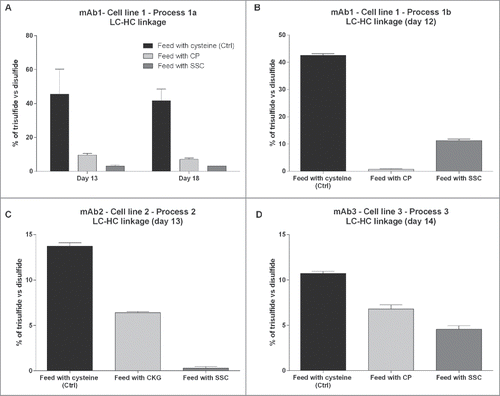
Table 1. Overview of the scale, culture conditions and processes of cell culture fed-batch experiments.
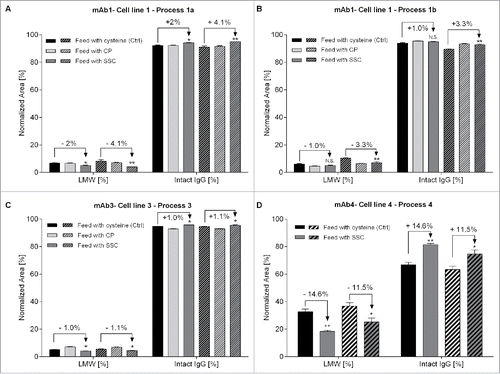
Figure 5. Relative quantification of intact IgG and total low molecular weight forms using capillary gel electrophoresis. Fragments were separated according to their size under non-reduced conditions using CE-SDS. (A) Quantification of mAb1 at day 13 (n = 6, plain bars) and day 18 (n = 6, hatched bars) of the fed-batch process 1a. (B) Quantification of mAb1 at day 5 (n = 8, plain bars) and day 9 (n = 8, hatched bars) of the fed-batch process 1b. (C) Quantification of mAb3 at day 14 (n = 8, plain bars) and day 18 (n = 8, hatched bars). (D) Quantification of mAb3 at day 8 (n = 6, plain bars) and day 12 (n = 6, hatched bars). Statistical differences were assessed by Kruskal-Wallis and Dunn's multiple comparison tests. P-values of less than 0.05 (*) and 0.01 (**) were considered significant. P-values higher than 0.05 are marked as non-significant (N.S.).