Figures & data
Figure 1. Schematic of stress study design. Humidity/thermal stress of infliximab samples were performed by incubating the drug powders at 40°C at different %RH for 0–4 weeks, followed by reconstitution in WFI and analysis.
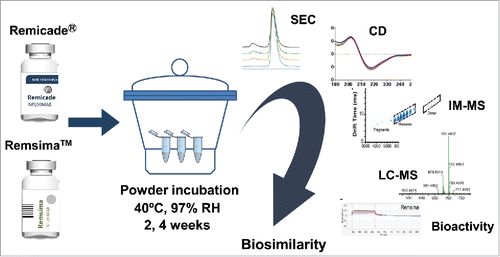
Figure 2. Characterization of protein aggregation in stressed samples. A. Representative SEC chromatograms of infliximab following 97% RH/40°C incubation for 0, 1, 2 and 4 weeks. B. SDS PAGE of Remicade® and Remsima™ samples stressed for 4 weeks at 0 (dry), 50, 75 and 97% RH. C. Kinetics of monomer loss at various humidity for Remicade® (solid) and Remsima™ (dashed) as detected by SEC (n = 4 ± SEM). D. Reducing SDS PAGE gel of Remicade® and Remsima™ humidity stressed samples for 4 weeks at 0 (dry), 50, 75 and 97% RH. Molecular weights (MW) are annotated on the ladder in kDa.
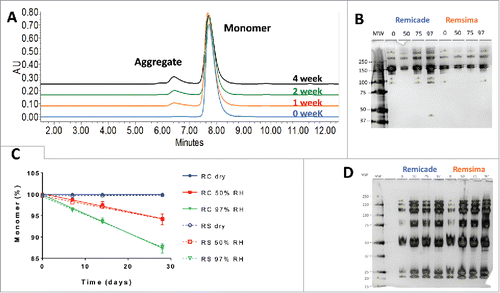
Table 1. Fitted rates of monomer loss for Remicade® and Remsima™ samples after incubation at 40°C and various relative humidity levels.
Figure 3. Nanoparticle tracking analysis of unstressed (light) and incubated at 97% RH/40°C for 4 weeks (dark) samples of Remicade® (A) and Remsima™ (B). DSC thermal melts of unstressed (dashed) and incubated at 97% RH/40°C for 4 weeks (solid) samples of Remicade® (C) and Remsima™ (D).
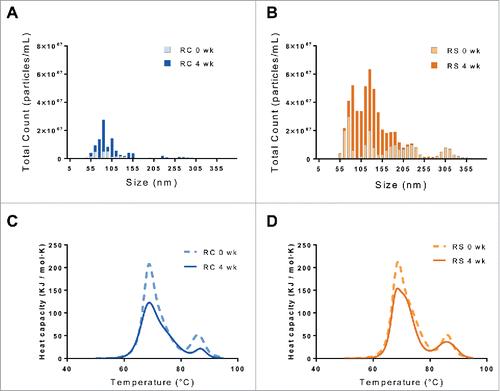
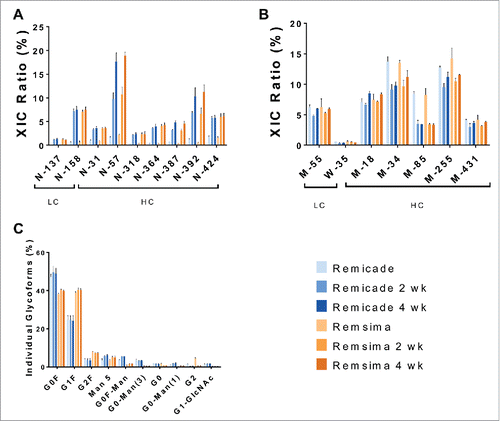
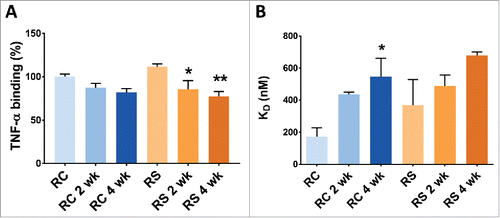
Table 2. Impurity profile of Remicade® and Remsima™ before and after 4-week incubation at 40°C and 97% relative humidity.
Figure 4. Ion mobility spectra and corresponding mass to charge spectrograms of Remicade® (A) and Remsima™ (B) before and after 4 weeks incubation at 97% RH/40°C, Remicade® (C) and Remsima™ (D). Fragment, monomer, dimer and trimer species annotated in ion mobility spectra.
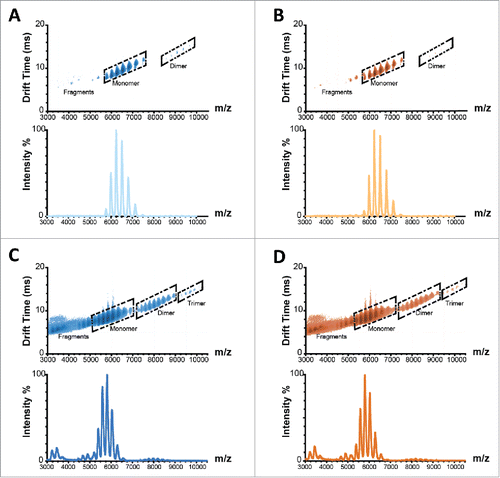
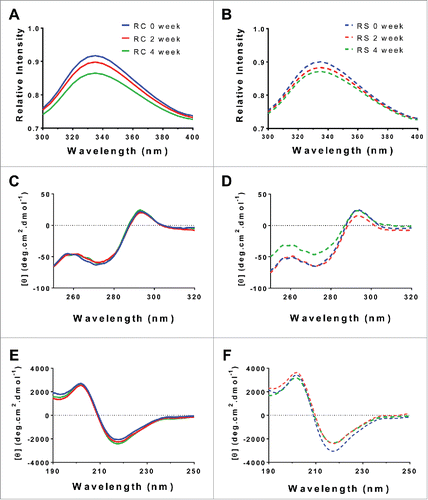
Figure 5. Biophysical characterization of humidity stressed samples of Remicade® (A, C, E, solid) and Remsima™ (B, D, F, dashed) incubated at 97% RH/40°C for 0 (blue), 2 (red) and 4 (green) weeks. Intrinsic fluorescence spectra ((A)and B), circular dichroism far UV spectra ((C)and D) and near UV spectra ((E)and F).