Figures & data
Figure 1. Isolation of nonspecific antibodies. A nonimmune scFv library was subjected to 4 rounds of yeast display-based selections. To ensure nonspecificity, the antigen used in each round was changed, with the following order used: HEK SMP, Sf9 SCP, Sf9 SMP, HEK SCP (Outlined boxes). The first 2 rounds were completed using magnetic-activated cell sorting and the final 2 rounds were completed using fluorescence-activated cell-sorting with the indicated sort gates (Thin boxes). The populations were validated after each round by measuring binding to each of the 4 reagents. As expected, the positively selected panel displays binding to all reagents.
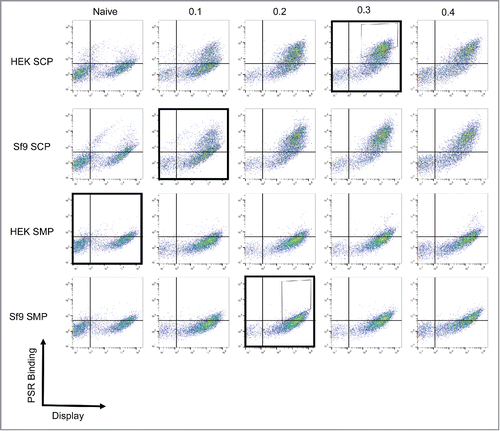
Figure 2. Antibody germline family distribution. Variable heavy (A) and light (B) chain sequences were aligned against human germline genes using IMGT V-QUEST and grouped into major families. Abundances of each family are displayed for the natural human repertoire (Blue), naive library (Red), negatively sorted pool (Green), and positively sorted population (Purple). Bars represent the average percentage of clones among 2 sorts and error bars indicate standard error.
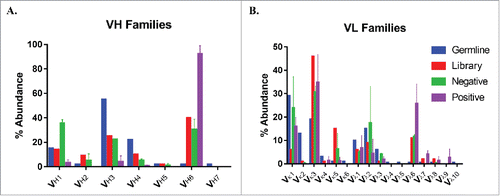
Figure 3. VH6 family sequence alignment. All VH6 antibody sequences were aligned using IMGT V-QUEST and sequence logos were plotted using Matlab for CDR H1 (A) and CDR H2 (B). Residues are numbered using Kabat standard numbering and CDRs are defined using IMGT definitions.Citation41
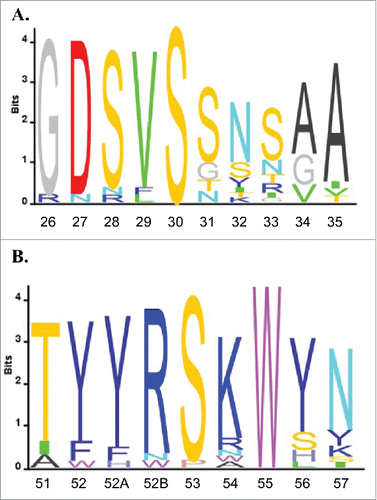
Figure 4. VH6 family CDRH2 mutations. One example VH6 clone was mutated by replacing CDRH2 with 2 different full H2 sequences from the VH1 family. PSR binding of the original clone (Blue) was compared against both mutants, VH6.H2.1 (Red) and VH6.H2.2 (Orange).
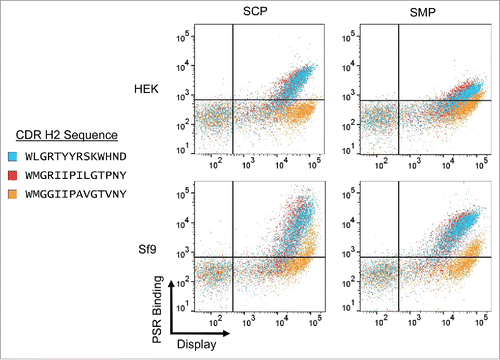
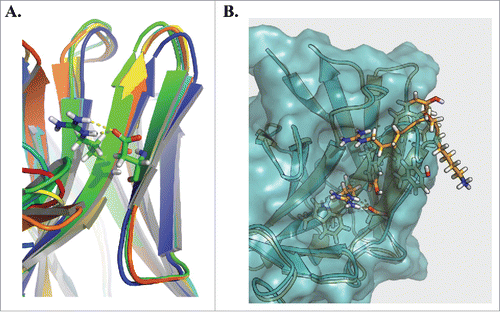
Figure 5. Homology modeling of the VH6 CDRH2. Rosetta Antibody was used to homology model 3 representative clones from the VH6 family (A). Alignment of the structures highlights a common constrained β sheet structure stabilized by hydrogen bond interactions between Arg50 and Asp58. Comparing this structure against that of clone VH6.H2.2 (B), we observe the CDRH2 of the VH6 family clone (orange) protrudes farther than that of the mutant (blue).