Figures & data
Figure 1. Major glycan structures of therapeutic mAbs. (a) Core-fucosylated agalacto-biantennary complex-type glycan (G0F); (b) core-fucosylated biantennary complex-type glycan with galactosylation on the Man α1-6 arm (G1aF); (c) core-fucosylated biantennary complex-type glycan with galactosylation on the Man α1-3 arm (G1bF); (d) core-fucosylated and fully galactosylated biantennary complex-type glycan (G2F). Yellow circle, Gal; green circle, Man; blue square, N-acetylglucosamine; red triangle, fucose.
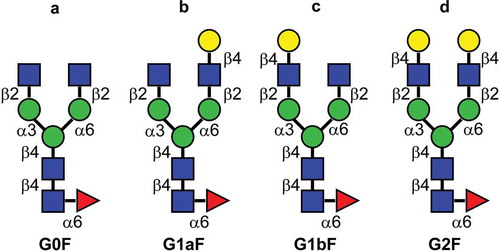
Figure 2. Preparation of anti-CD20 mAbs with core-fucosylated homogeneous N-glycans. This illustration details the preparation of anti-CD20 mAbs having two N-glycan chains with G2F structure. (1) digestion with wild-type Endo S and Endo D for truncating N-glycans, (2) transglycosylation with Endo F3 mutant (D165Q) in the presence of oxazolinated glycans as a glycan donor, (3) the separation and purification of anti-CD20 mAbs with two core-fucosylated N-glycan chains using cation-exchange column chromatography. Yellow circle, Gal; green circle, Man; blue square, N-acetylglucosamine; red triangle, fucose.
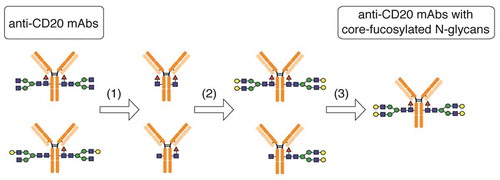
Table 1. Fc-glycopeptides analysis of mAbs with homogenous glycan.
Figure 3. Glycan profiles of mAbs with homogeneous glycans. Glycan profiles of commercially available anti-CD20 mAb (a) and glycoengineered G0F mAb (b), G1aF mAb (c), G1bF mAb (d), and G2F mAb (e). The glycan profiles were obtained using HPLC analysis of 2-AB-labeled glycans.
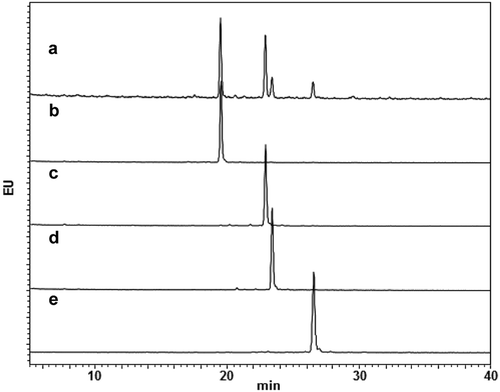
Figure 4. CDC and C1q-binding activities of glycoengineered anti-CD20 mAbs. (a, b) CDC activities of glycoengineered anti-CD20 mAbs. Raji cells were incubated with 16% human serum and were serially diluted with glycoengineered anti-CD20 mAbs. (a) Percentages of cell lysis plotted against mAb concentrations. (b) CDC activity of 1 µg/ml of G1aF mAb and G1bF mAb mixtures in different ratios. (c) C1q binding of glycoengineered anti-CD20 mAbs. Raji cells were opsonized with anti-CD20 mAbs and incubated with human serum. The cells were stained with FITC-conjugated anti-C1q antibody and the C1q-binding level was analyzed by flow cytometer. Data are presented as mean ± SD (n = 3).
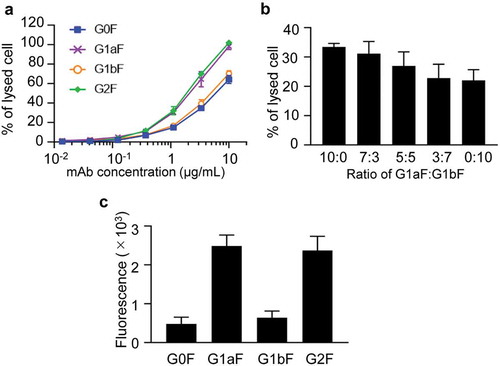
Figure 5. Binding of glycoengineered anti-CD20 mAbs to human FcγRs. SPR analysis was used to measure the binding of anti-CD20 mAbs to human FcγRI, FcγRIIa, and FcγRIIIa. Binding sensorgrams corrected by both the surface blank and buffer injection control are represented.
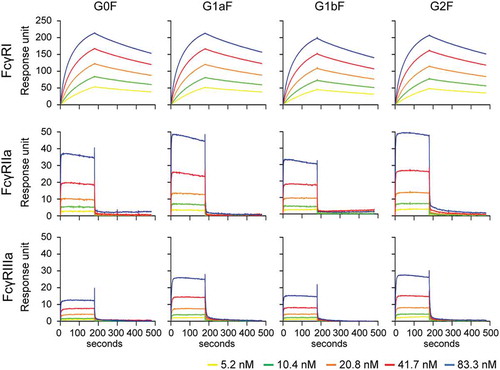
Figure 6. FcγR activation properties of glycoengineered anti-CD20 mAbs. FcγRIIa (a) and FcγRIIIa (b) activation properties of glycoengineered mAbs. Jurkat/FcγRIIa/NFAT-Luc or Jurkat/FcγRIIIa/NFAT-Luc reporter cells were incubated with serially diluted anti-CD20 mAbs in the presence of Raji cells. FcγR activation was evaluated by assessing the luminescence intensity.
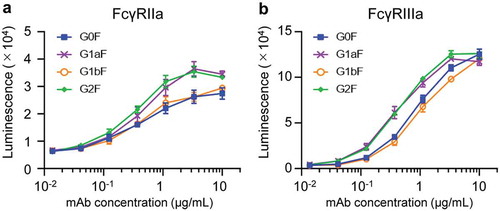
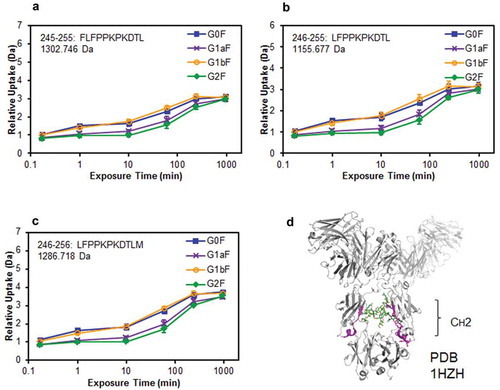
Figure 7. Comparison of structural stabilities of the CH2 domain of glycoengineered anti-CD20 mAbs using HDX/MS. Deuterium uptake plots of peptides Phe245–Leu255 (FLFPPKPKDTL) (a), Leu246–Leu255 (LFPPKPKDTL) (b), and Leu246–Met256 (LFPPKPKDTLM) (c) in the H-chain. (d) Physical representations of the crystal structures (PDB 1HZH) of peptides at Phe245–Met256 (magenta ribbons).