Figures & data
Figure 1. Properties of the Li81 Fab–LINGO-1 ectodomain complex. Binding interfaces of the Li81 Fab-LINGO-1 ectodomain complex determined from the crystal structure.Citation20 Structural figures were rendered with MOE software.Citation23 (a) Contacts comprising the primary binding interface, between Li81 (pink) CDR residues and LINGO-1 (green) LRR domains 4–8. (b) Contacts comprising both the primary interface and the secondary binding interface, between Li81 (pink) light chain framework residues and the Ig domain of a separate molecule of LINGO-1 (yellow)
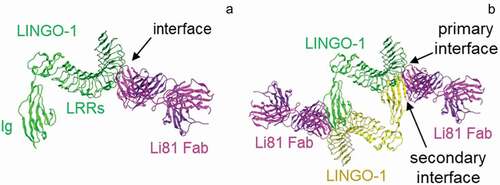
Figure 2. Concentration-dependent oligomerization of Li81 mAb-LINGO-1 ectodomain complexes. Samples containing 10 µg LINGO-1 ectodomain and 0, 3 10, or 30 µg of Li81 mAb, or 10 µg Li81 mAb alone, were subjected to analytical SEC. The column effluent was analyzed for absorbance at 280 nm and in-line light scattering. (a) Chromatograms from the SEC column. Y-axis: milli-absorbance units (mAU). X-axis: time in minutes after injection. (b) Image of three representative 34 nm particles observed by EM from a sample containing 10 µg each of Li81 mAb and LINGO-1 ectodomain and model of the 34 nm particle. (c and d) SDS-PAGE/Western blot analysis of LINGO-1 in CHO cells expressing HA LINGO-1 following treatments for 2 days with serial dilutions of the Li81 mAb (c) and Li81 Fab (d). Apparent molecular weights in kiloDaltons (kDa), based on the positions of pre-stained markers, are shown at the left
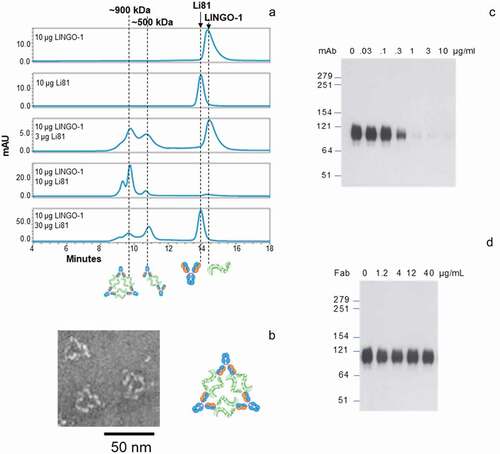
Figure 3. The secondary LINGO-1 binding site of Li81 mAb. (a) Interfaces between Li81 and LINGO-1 LRR domains and the Ig domain of the second LINGO-1 molecule identified in the pdb:4OQT crystal structure, with the three mutations in CN1373 and the glycan on LINGO-1 N225 highlighted. (b) Close-up of Li81 VL R18 and LINGO-1 Ig domain contacts, with distances for the closest contacts shown in green (Å). (c) Close-up of Li81 VL R54 and LINGO-1 Ig domain contacts, and possible hydrogen bonds to backbone O atoms in each molecule. (d) Close-up of where Li81 VL S31 could, if mutated, contact the glycan at N225 of LINGO-1 LRR region and the second LINGO-1 molecule Ig domain. (e) Sequence of Li81 VL. Li81 residues with lines above indicate the primary binding site (i.e., interatomic distance within 4.5 Å of LINGO-1 LRR domains); lines below indicate the secondary binding site (i.e., within 4.5 Å of LINGO-1 Ig domain). Li113 VL, which inspired most of the mutations, is shown for comparison; S31R and 4 other differences in the secondary binding site are emboldened and highlighted in yellow. CDRs are highlighted in gray
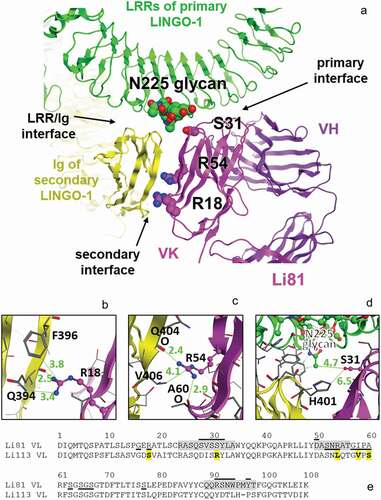
Table 1. Thermal stability and binding affinity of Li81 light chain variant antibodies designed to reduce binding at the secondary site. Tm values were measured by differential scanning fluorimetry. EC50 values were calculated in an ELISA study assessing binding on LINGO-1-Fc coated plates
Figure 4. Biochemical attributes of Li81 variants targeting secondary binding site contacts. (a) Samples (4 µg/lane) were subjected to SDS-PAGE and stained with Coomassie brilliant blue. Samples on the left were analyzed under reducing conditions, and samples on the right were analyzed under non-reducing conditions. Molecular weight markers and their apparent molecular masses are shown at the left of the panel. (b) The apparent affinities of the seven secondary binding site mutants for LINGO-1 were measured by a direct-binding ELISA. Data are plotted as absorbance at 450 nm versus concentration. (c) Samples, each containing 10 µg LINGO-1 ectodomain and 10 µg of of one of the mutant mAbs, were subjected to SEC on an analytical SEC column using PBS as the mobile phase. The column effluent was analyzed for absorbance at 280 nm. (d) Samples containing CN1373 mAb alone, or 3 µg LINGO-1 ectodomain and 1.6, 3, or 10 µg of CN1373 mAb, were subjected to SEC with in-line light scattering
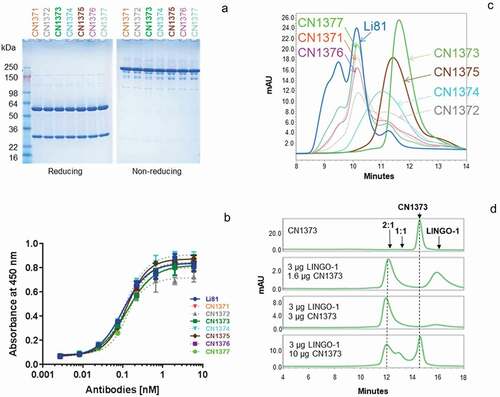
Figure 5. Activity of the Li81 secondary binding site mutants in the OPC differentiation and coculture assays. In A and B, samples were evaluated in the OPC differentiation bioassay using expression of MBP as a readout for differentiation. MBP expression was quantified by ICC and recorded as MFI per differentiated oligodendrocyte (Olig2+ cell). (a) Dose-response of Li81, CN1373, and CN1375 in the OPC differentiation assay. Data shown are from 2 independent studies. (b) Activity of Li81, LINGO-1, CN1373, and premixed CN1373/LINGO-1 complex in the OPC differentiation assay. C. DRG neuron/A2B5+ cell cocultures were treated with 3 µg/mL of Li81, CN1373, or 5C8 isotype control antibody. The cultures were fixed on day 10 and visually analyzed for axonal myelination by oligodendrocytes by ICC using anti-MBP (green, 488) and anti-tubulin (red, 594) antibodies for detection of myelinating oligodendrocytes and neurons, respectively
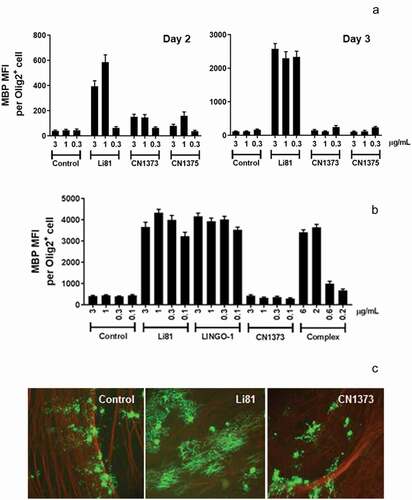
Figure 6. Internalization of LINGO-1 following treatment with Li81 and CN1373. (a) CHO cells expressing full-length LINGO-1 were treated with 5C8 isotype control antibody (Ctl), Li81, or CN1373 at 1 and 3 µg/mL for 23 h at 37°C and analyzed by FACS, staining for anti-human IgG. (b) Ba/F3 cells expressing full-length LINGO-1 were treated for 3 h at 3°C or 37°C with serial dilutions of 5C8, Li81, or CN1373 and analyzed by FACS. (c) Rat primary cortical neurons cultured for 12 days were treated with isotype control (5C8) and LINGO-1 specific antibodies for 90 min at 37°C, acid washed and fixed for with 1.5% paraformaldehyde for 5 min at room temperature. For assessing intracellular trafficking, the cells were treated with detergent, and then treated with Alexa-647 secondary antibody (red) for detection of Li81, CN1373, and 5C8, and Alexa-488 (green) for the lysosomal marker Rab7. Representative confocal middle sections of each condition are shown. Selected regions (stippled-line squares) were cropped and enlarged and are displayed using an inverted monochrome color scale to aid visualization. Scale bar, 5 µm
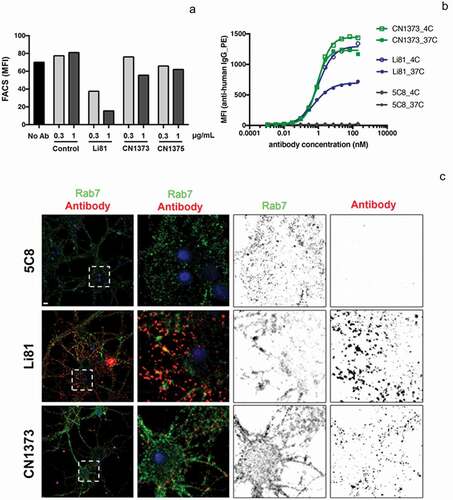
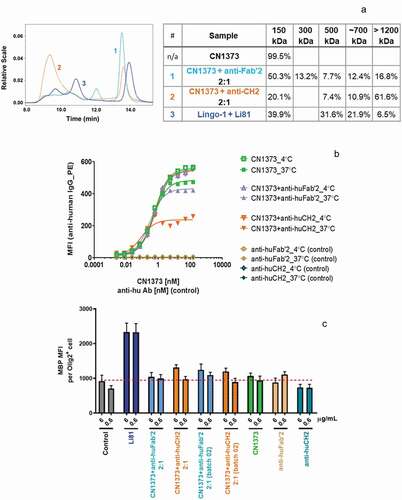
Figure 7. Internalization of LINGO-1 following treatment with complex forms of CN1373. (a) SEC chromatograms and tabulated summary of the data from samples containing CN1373 that were clustered with anti-human CH2 or Fab′2 antibody. (b) Ba/F3 cells expressing full length LINGO-1 were treated for 3 h at 4°C or 37°C with serial dilutions of CN1373, anti-CH2-1373 complexes, and anti-Fab′2-1373 complexes and analyzed by FACS. (c) Activity of CN1373, anti-CH2-1373 complexes, and anti-Fab′2-1373 complexes in the OPC differentiation assay. MBP expression was quantified by ICC and recorded as MFI per differentiated oligodendrocyte (Olig2+ cell)