Figures & data
Figure 1. Competitive binding between anti-OVA antibodies. Capture and probe antibodies refer to antibodies in solid phase used to capture OVA and antibodies in solution phase in the assay used to determine cross-competition, respectively. Red and red/black cross-hatched squares indicate no binding of the probe antibody. Green squares indicate binding of the probe antibody. Yellow squares indicate weak binding of probe antibody. OVA-36 and 46, highlighted in black, share IGHV5-25 and IGKV5S2 germline segment use but have distinct binding competition patterns. Groups of squares highlighted with white lines indicate competition of clones with the same VH/VL germline segment pairing. Names of antibodies within each VH/VL germline segment group are color-coded as in Suppl. Figure 2. Only antibodies with competition data as both capture and probe reagents as well as OVA-36 and 46 are shown, with the full dataset shown in Suppl. Figure 2
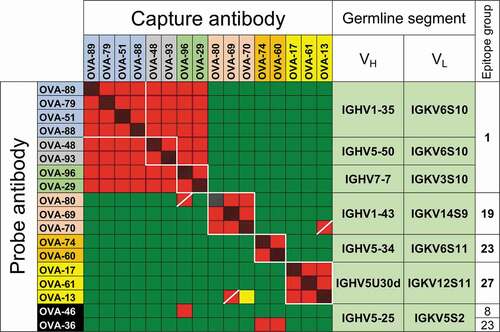
Figure 2. VH and VL germline segments and CDR H3 sequences of anti-OVA clones in parallel lineages. Amino acid identities between 2 or more clones within the same parallel lineage with the same CDR H3 length are shown in black and gray backgrounds. Randomly selected clones that were and were not part of parallel lineages in the initial antibody panel are highlighted in orange and yellow, respectively. Clones selected based on VH/VL germline segment pair use are highlighted in blue
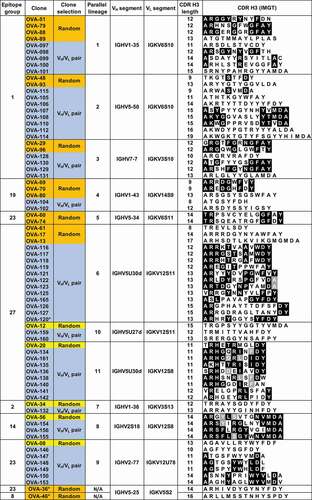
Figure 3. Binding competitions between anti-OVA antibodies in parallel lineages. Red and red/black cross-hatched squares indicate no binding of the probe antibody. Green squares indicate binding of the probe antibody. Yellow and gray squares indicate weak and indeterminate binding of probe antibody, respectively. Squares with diagonal bars indicate discordant one-way competition results within parallel lineages. Clones are highlighted in orange, yellow and blue as in . Clones with white background are not part of parallel lineages and were used to discriminate between epitope groups. Clone OVA-164 is a clonal variant of OVA-64. Competition patterns for reference antibodies of parallel lineages 8, 9 and 10 are shown in Suppl. Fig. 6B
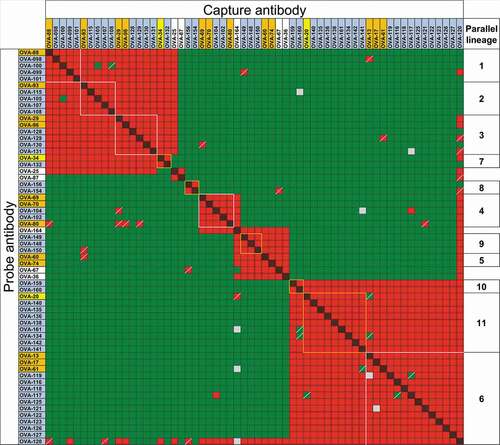
Figure 4. Epitope mapping of anti-HEL antibodies. (a) Competition patterns of anti-HEL clones tested. Red and red/black cross-hatched squares indicate no binding of the probe antibody. Green squares indicate binding of the probe antibody. Mouse antibodies of known epitope specificity are highlighted in shades of blue, orange, red and purple. Anti-HEL clones selected based on VH/VL pairing are shown with asterisks. White lines and dotted lines indicate cross-competition with other clones in parallel lineages and mouse anti-HEL antibodies. (b) Composite structure of HEL bound to reference mouse antibodies (PDB 1YQV, 1FDL, 1FBI, 1MLC, 1NDG and 1JHL). Variable regions of antibodies are color-coded as in panel a. (c) Graphical representation of epitope groups of antibodies in parallel lineages, mouse anti-HEL antibodies and additional selected rat anti-HEL antibodies. Antibodies and parallel lineages colored as in panels a and b
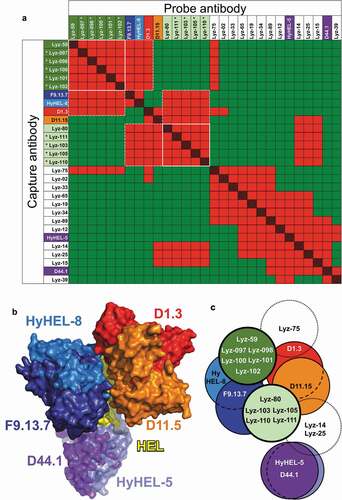
Figure 5. CDR H3 consensus sequences of unique anti-Her2 hu4D5 (a) and hu2C4 (b) variant clones. Parental CDR H3 sequence regions randomized in libraries are shown above the sequence logos, highlighting residues contacting Her2 through side-chains in red and through main-chain in blue. The number of unique clones for each group are shown
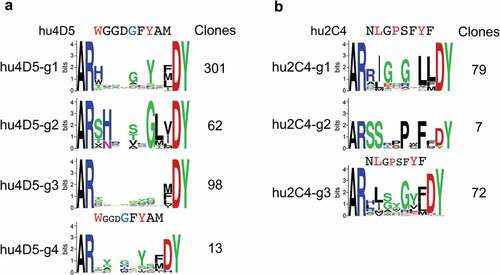
Table 1. CDR H3 sequence and binding kinetics of best anti-Her2 variants in different consensus groups
