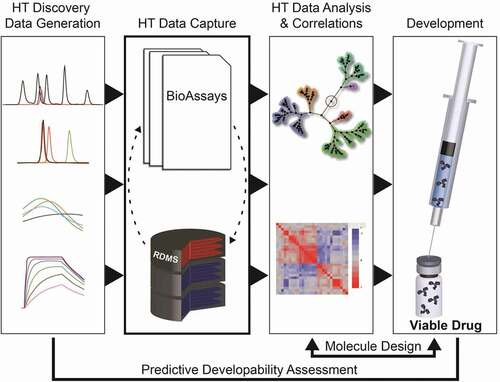
Figures & data
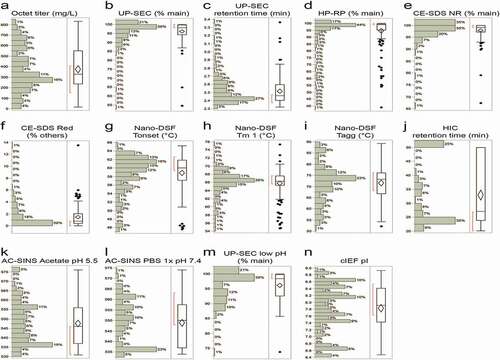
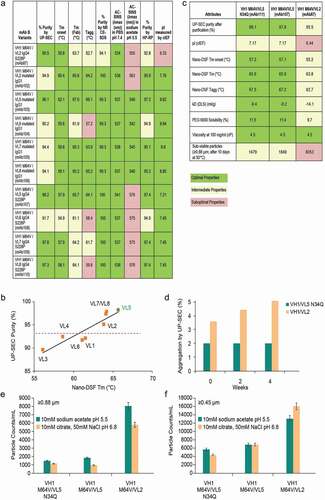
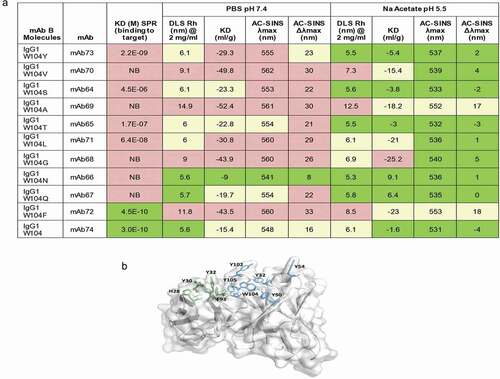
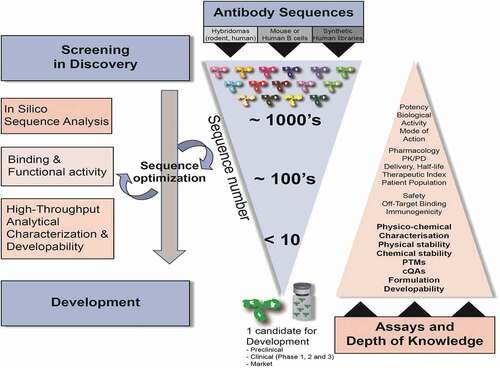
Table 1. Critical molecule properties and analytical assays used during sequence selection and developability assessment
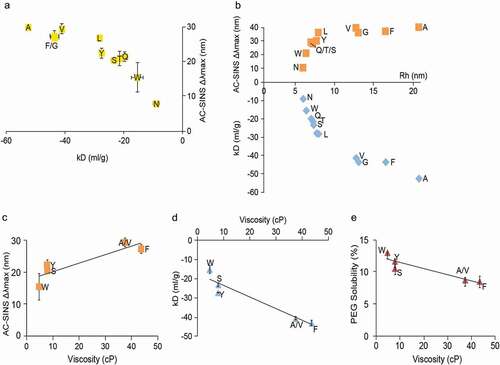
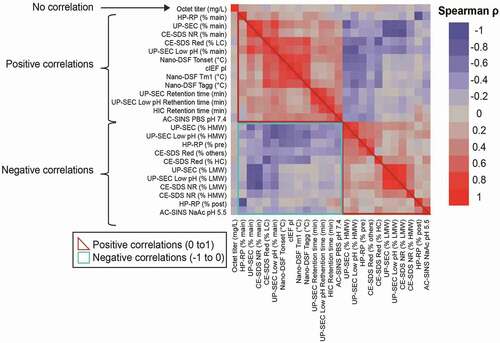
Table 2. Spearman correlations (ρ > 0.5) for selected analytical characterization read-outs with p-values <0.0001. Pearson coefficients and associated p-values are also shown. P-values test null hypothesis that the correlation coefficient = 0
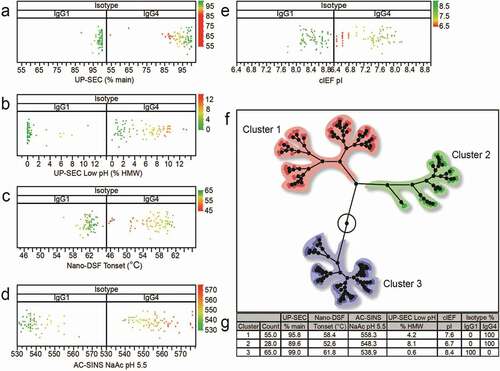
Table 3. Spearman correlations (ρ > 0.5) for selected analytical characterization read-outs with p-values <0.0001 separated by isotype (IgG1 and IgG4). Pearson coefficients and associated p-values are also shown. P-values test null hypothesis that the correlation coefficient = 0
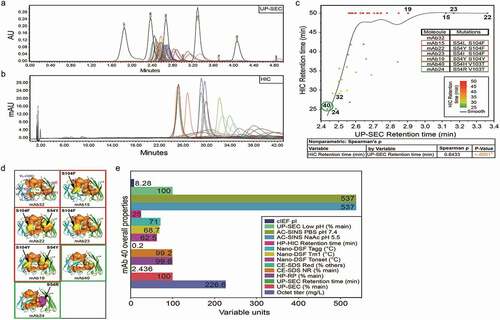
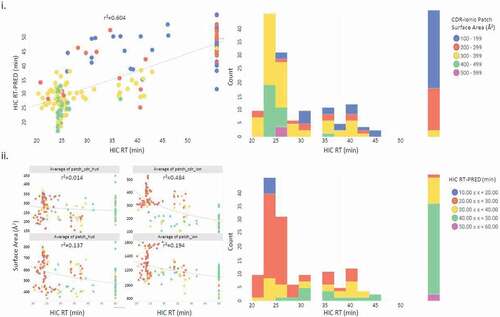
Table 4. Experimental and predicted HIC retention times for selected mAbs. Included are the 4 contributing properties which compose the 4-Pt QSPR equation resulting in the predicted retention times. r2 values for each column vs. HIC RT are displayed in the final row
Wang W. Protein aggregation and its inhibition in biopharmaceutics. Int J Pharm. 2005;289:1–30. doi:https://doi.org/10.1016/j.ijpharm.2004.11.014. Cromwell MEM, Hilario E, Jacobson F. Protein aggregation and bioprocessing. Aaps J. 2006 Sep 15;8(3):E572–9. Vlasak J, Ionescu R. Heterogeneity of monoclonal antibodies revealed by charge-sensitive methods. Curr Pharm Biotechnol. 2008;9:468–81. doi:https://doi.org/10.2174/138920108786786402. Liu H, Gaza-Bulseco G, Faldu D, Chumsae C, Sun J. Heterogeneity of monoclonal antibodies. J Pharm Sci. 2008;97:2426–47. doi:https://doi.org/10.1002/jps.21180. Hanke AT, Klijn ME, Verhaert PDEM, van der Wielen LAM, Ottens M, Eppink MHM. van de Sandt EJAX. Prediction of protein retention times in hydrophobic interaction chromatography by robust statistical characterization of their atomic-level surface properties. Biotechnol Prog. 2016;32:372–81. doi:https://doi.org/10.1002/btpr.2219. Hebditch M, Roche A, Curtis RA, Warwicker J. Models for antibody behavior in hydrophobic interaction chromatography and in self-association. J Pharm Sci. 2019;108:1434–41. doi:https://doi.org/10.1016/j.xphs.2018.11.035. Kelly RL, Sun T, Jain T, Caffry I, Yu Y, Cao Y, Lynaugh H, Brown M, Vásquez M, Dane Wittrup K, et al. High throughput cross-interaction measures for human IgG1 antibodies correlate with clearance rates in mice. MAbs. 2015;7:770–77. doi:https://doi.org/10.1080/19420862.2015.1043503. Jacobitz AW, Dykstra AB, Spahr C, Agrawal NJ. Effects of buffer composition on site-specific glycation of lysine residues in monoclonal antibodies. J Pharm Sci. 2019 Jan;109(1):293–300. doi: https://doi.org/10.1016/j.xphs.2019.05.025. Epub 2019 May 29. Goyon A, Excoffier M, Janin-Bussat MC, Bobaly B, Fekete S, Guillarme D, Beck A. Determination of isoelectric points and relative charge variants of 23 therapeutic monoclonal antibodies. J Chromatogr B Anal Technol Biomed Life Sci. 2017;1065–1066:119–28. doi:https://doi.org/10.1016/j.jchromb.2017.09.033. Breitsprecher D, Glücklich N, Hawe A, Menzen T Comparison of nanoDSF and µDSC for thermal stability assessment during biopharmaceutical formulation development [Internet]. 2006 [cited 2020 Jan 27]; Available from: https://resources.nanotempertech.com/application-notes/application-note-nt-pr-006-dsc-nanodsf-comparison Estep P, Caffry I, Yu Y, Sun T, Cao Y, Lynaugh H, Jain T, Vásquez M, Tessier PM, Xu Y. An alternative assay to hydrophobic interaction chromatography for high-throughput characterization of monoclonal antibodies. MAbs. 2015;7:553–61. doi:https://doi.org/10.1080/19420862.2015.1016694. Dani B, Platz R, Tzannis ST. High concentration formulation feasibility of human immunoglobulin G for subcutaneous administration. J Pharm Sci. 2007;96:1504–17. doi:https://doi.org/10.1002/jps.20508. Hofmann M, Winzer M, Weber C, Gieseler H. Limitations of polyethylene glycol-induced precipitation as predictive tool for protein solubility during formulation development. J Pharm Pharmacol. 2018;70:648–54. doi:https://doi.org/10.1111/jphp.12699. Sankar K, Hoi KH, Yin Y, Ramachandran P, Andersen N, Hilderbrand A, McDonald P, Spiess C, Zhang Q. Prediction of methionine oxidation risk in monoclonal antibodies using a machine learning method. MAbs. 2018;10:1281–90. doi:https://doi.org/10.1080/19420862.2018.1518887. Lu X, Nobrega RP, Lynaugh H, Jain T, Barlow K, Boland T, Sivasubramanian A, Vásquez M, Xu Y. Deamidation and isomerization liability analysis of 131 clinical-stage antibodies. MAbs. 2019;11:45–57. doi:https://doi.org/10.1080/19420862.2018.1548233. Robinson NE, Robinson AB. Prediction of protein deamidation rates from primary and three-dimensional structure. Proc Natl Acad Sci U S A. 2001;98:4367–72. doi:https://doi.org/10.1073/pnas.071066498. Hao P, Adav SS, Gallart-Palau X, Sze SK. Recent advances in mass spectrometric analysis of protein deamidation. Mass Spectrom Rev. 2017 Nov;36(6):677–92. Epub 2016 Jan 13. doi:https://doi.org/10.1002/mas.21491. Lechner A, Giorgetti J, Gahoual R, Beck A, Leize-Wagner E, François YN. Insights from capillary electrophoresis approaches for characterization of monoclonal antibodies and antibody drug conjugates in the period 2016–2018. J Chromatogr B Anal Technol Biomed Life Sci. 2019;1122–1123:1–17. doi:https://doi.org/10.1016/j.jchromb.2019.05.014. Azadi P, Heiss C. Mass spectrometry of N-linked glycans. Methods Mol Biol. 2009;534:37–51. doi:https://doi.org/10.1007/978-1-59745-022-5_3. Miyanabe K, Yamashita T, Abe Y, Akiba H, Takamatsu Y, Nakakido M, Hamakubo T, Ueda T, Caaveiro JMM, Tsumoto K. Tyrosine sulfation restricts the conformational ensemble of a flexible peptide, strengthening the binding affinity for an antibody. Biochemistry. 2018;57:4177–85. doi:https://doi.org/10.1021/acs.biochem.8b00592. Banks DD, Gadgil HS, Pipes GD, Bondarenko PV, Hobbs V, Scavezze JL, Kim J, Jiang XR, Mukku V, Dillon TM. Removal of cysteinylation from an unpaired sulfhydryl in the variable region of a recombinant monoclonal IgG1 antibody improves homogeneity, stability, and biological activity. J Pharm Sci. 2008;97:775–90. doi:https://doi.org/10.1002/jps.21014. Mason M, Sweeney B, Cain K, Stephens P, Sharfstein ST. Identifying bottlenecks in transient and stable production of recombinant monoclonal-antibody sequence variants in chinese hamster ovary cells. Biotechnol Prog. 2012;28:846–55. doi:https://doi.org/10.1002/btpr.1542. Benner SW, Welsh JP, Rauscher MA, Pollard JM. Prediction of lab and manufacturing scale chromatography performance using mini-columns and mechanistic modeling. J Chromatogr A. 2019;1593:54–62. doi:https://doi.org/10.1016/j.chroma.2019.01.063. He F, Woods CE, Trilisky E, Bower KM, Litowski JR, Kerwin BA, Becker GW, Narhi LO, Razinkov VI. Screening of monoclonal antibody formulations based on high-throughput thermostability and viscosity measurements: design of experiment and statistical analysis. J Pharm Sci. 2011;100:1330–40. doi:https://doi.org/10.1002/jps.22384. Alfthan K. Surface plasmon resonance biosensors as a tool in antibody engineering. In: biosensors and Bioelectronics. Elsevier Sci Ltd. 1998 Sep 15;13(6):653–63. Xu Y, Roach W, Sun T, Jain T, Prinz B, Yu TY, Torrey J, Thomas J, Bobrowicz P, Vásquez M, et al. Addressing polyspecificity of antibodies selected from an in vitro yeast presentation system: A FACS-based, high-throughput selection and analytical tool. Protein Eng Des Sel. 2013;26:663–70. doi:https://doi.org/10.1093/protein/gzt047. Avery LB, Wade J, Wang M, Tam A, King A, Piche-Nicholas N, Kavosi MS, Penn S, Cirelli D, Kurz JC, et al. Establishing in vitro in vivo correlations to screen monoclonal antibodies for physicochemical properties related to favorable human pharmacokinetics. MAbs. 2018;10:244–55. doi:https://doi.org/10.1080/19420862.2017.1417718. Goulet DR, Atkins WM. Considerations for the design of antibody-based therapeutics. J Pharm Sci. 2019. Beck A, Wurch T, Bailly C, Corvaia N. Strategies and challenges for the next generation of therapeutic antibodies. Nat Rev Immunol. 2010;10:345–52. doi:https://doi.org/10.1038/nri2747.