Figures & data
Table 1. Binding and functional properties of hits derived from the full naïve campaign
Figure 1. (A). Scouting campaign selection strategy. In order to identify a single antigen format to take forward into a naïve campaign applying the full AdimabTM Library diversity, a subset of the library was subjected to multiple rounds of selection using biotinylated CHO-hCCR1 cells, hCCR1-VLPs, or combinations thereof. Some samples were additionally subjected to pre-clearing with control cells using MACS® prior to positive selection with the antigen as shown. Following selection round 4, clones from each output were generated as yeast supernatants and screened for hCCR1 binding, and specificity for hCCR1 vs hCCR5, on appropriate recombinant cell lines. (B). Characterization of scouting campaign output. Clones selected from the ‘scouting’ campaign, as outlined in Figure 1a, were screened using a homogenous cell binding assay (FMAT) to assess binding to CHO-hCCR1, and specificity for hCCR1 by comparing binding to CHO-hCCR1 and CHO-hCCR5 cells. Each full-length mAb clone was expressed in yeast supernatant; duplicate data points were averaged and the CHO-hCCR1:CHO-hCCR5 binding ratio calculated. A cutoff binding ratio of >2 was applied and the resulting dataset grouped depending on whether individual clones were derived from selections using CHO-hCCR1 cells only, hCCR1-VLPs only, or combinations of both antigens
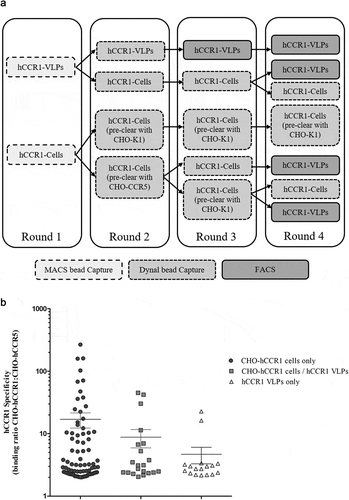
Figure 2. (A). Full naïve campaign selection strategy. The full AdimabTM library was subjected to four rounds of MACS® selections on biotinylated CHO-hCCR1 cells. At rounds 2 to 4, a preclear was performed using biotinylated CHO-hCCR5 or CHO-K1 (parental) cells. Following selection round 4, individual clones were screened as yeast supernatants using a homogenous cell binding assay (FMAT) to assess binding to CHO-hCCR1, and specificity for hCCR1 by comparing binding to CHO-hCCR1 and CHO-hCCR5 cells. (B). Full naïve campaign binding screen. Clones selected from the full naïve campaign, as outlined in (A), were assessed for binding to CHO-hCCR1 and -hCCR5 in a homogenous cell binding assay (FMAT). Each full-length mAb clone was tested in ‘single-shot’ over two independent assay runs. The data shown represents mean FMAT signal values ± SEM across the two runs for clones with a binding ratio (CHO-hCCR1:CHO-hCCR5) >20; these data were ordered from left to right (lowest to highest averaged FMAT signal for CHO-hCCR1)
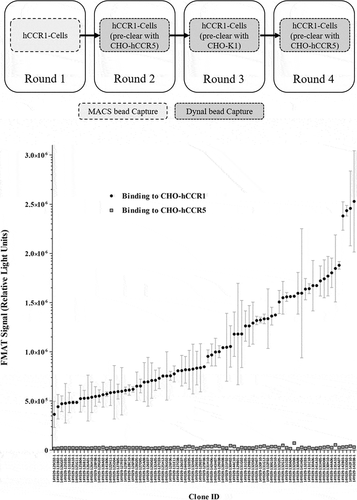
Figure 3. Inhibition of RANTES/CCL5-induced human donor monocyte chemotaxis. CD14+ monocytes were isolated from human PBMCs using MACS® technology, and chemotaxis of monocytes toward RANTES/CCL5 assessed in a transwell assay format. Monocytes were pre-incubated with mAb 14Y029-133D02 prior to addition of the chemokine. An IC50 value of 371 pM was calculated in this example dataset generated with monocytes derived from a specific human donor. The full dataset for 14Y029-133D02 using monocytes derived from multiple donors is shown in.
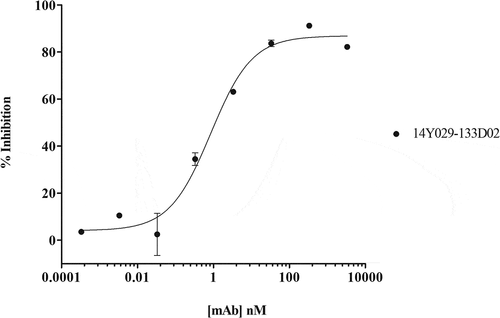
Figure 4. (A). QC of purified his-tagged hCCR1. His-tagged hCCR1 was expressed in a stable HEK293 cell line. Cells were disrupted and purified membranes solubilized with DDM/CHS; hCCR1-his was subsequently captured from the supernatant on TALON® immobilized metal affinity chromatography (IMAC) resin and eluted with 250 mM imidazole. Monodispersity was assessed using analytical size-exclusion chromatography coupled to tryptophan fluorescence detection, while purity was estimated by SDS-PAGE followed by Coomassie dye staining (inset). (B). Affinity maturation campaign selection strategy. Maturation libraries were subjected to one round of magnetic bead selection and 3 rounds of FACS selections. At round 3 a null selection was performed using a polyspecificity reagent (PSR)Citation41 designed to remove cross-reactive and sticky clones. At round 4 the antigen concentration was reduced to 1 µg/ml to aid identification of clones with improved affinity. Following the round 4 selection individual cones were evaluated for binding to CHO-hCCR1 cells. (C). Affinity-maturation of anti-CCR1 mAb clone 14Y029-142B03 using purified hCCR1 as antigen. Affinity-matured variants of parental mAb 14Y029-142B03, derived from the full naïve library selections, were isolated from diversified in CDRH1 and CDRH2 libraries using purified hCCR1 as antigen. Binding of test mAbs to CHO-hCCR1 cells was determined by flow cytometry via a fluorescently labeled detection antibody; a representative EC50 curve for the ‘most improved’ variant of parental mAb 14Y029-142B03 is shown
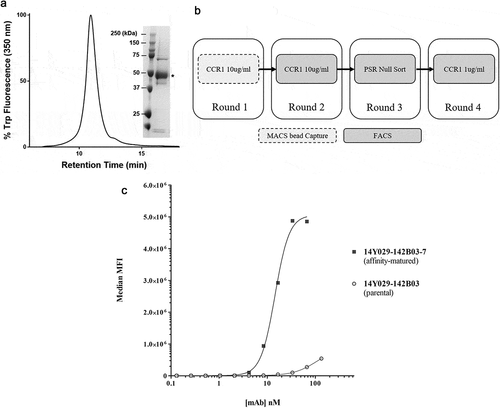
Table 2. Inhibition of RANTES/CCL5-induced human donor monocyte chemotaxis
