Figures & data
Figure 1. Broadly neutralizing antibodies against HIV-1 generally have substantial somatic mutations and often have Fv N-linked glycosylation. (a) Sequences of glycosylated Fv heavy chain (HC) or light chain (LC) for four HIV-1 broadly neutralizing antibodies. Somatic hypermutations are shown, with introduced N-linked glycan sequon labeled with green circles. (b) Structures of the four glycosylated antigen-binding fragments (Fabs). Glycans of occupied N-linked glycosylation sequon are displayed on structures of PGT121 (PDB:4FQ1), CAP256-VRC26.25 (PDB:5DT1), N6 (PDB:5TE6), and VRC07-523 (modeled from PDB:4OLW) as shown in green sticks, and with the most frequent glycoform being shown (see Table S1). The two N-linked glycosylation sequon on PGT121 that were not glycosylated (N58 and N69, see Table S2 and S3) were shown in green spheres
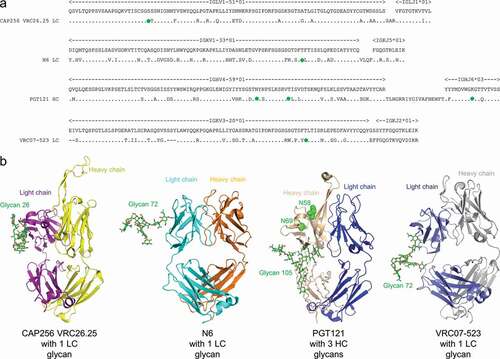
Figure 2. Polyreactivity of N6 and VRC07-523LS is enhanced by reversion of somatic mutation, N72LCT, which removes the introduced N-linked glycan. (a) HEp-2 cell staining (antibody concentration: 25 µg/ml) and (b) anticardiolipin ELISA for CAP256-VRC26.25, N6, PGT121, VRC07-523LS, and their glycan knockouts based on germline encoding residues. For CAP256-VRC26.25 and PGT121 there was no observable difference between wild type and glycan knockouts in both assays, while for N6 and VRC07-523LS higher polyreactivity for the glycan knockout was observed
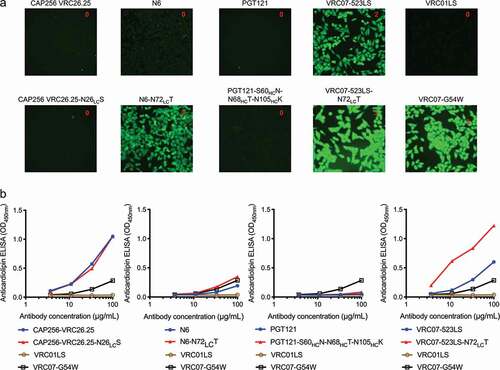
Figure 3. Neutralization, thermostability, pharmacokinetics, and heterogeneity of CAP256-VRC26.25-N26LCS. (a) Neutralization IC50 (µg/ml) of CAP256-VRC26.25 wild type and CAP256-VRC26.25-N26LCS assessed with nine HIV-1 strains. (b) Pharmacokinetic profile for CAP256-VRC26.25 wild type and CAP256-VRC26.25-N26LCS in humanized FcRn mice. Dash line denotes limit of detection. (c) Extracted ion chromatogram (XIC) of [366.137 ± 0.0005] Da corresponding to the signature glycan peak in the trypic digest of CAP256-VRC26.25 samples: a distribution of the light chain (Fv) glycopeptides in the control material (top trace); only the heavy chain (Fc) glycopeptides are observed in the CAP256-VRC26.25-N26LCS material (bottom trace). XIC peak intensities have been normalized
![Figure 3. Neutralization, thermostability, pharmacokinetics, and heterogeneity of CAP256-VRC26.25-N26LCS. (a) Neutralization IC50 (µg/ml) of CAP256-VRC26.25 wild type and CAP256-VRC26.25-N26LCS assessed with nine HIV-1 strains. (b) Pharmacokinetic profile for CAP256-VRC26.25 wild type and CAP256-VRC26.25-N26LCS in humanized FcRn mice. Dash line denotes limit of detection. (c) Extracted ion chromatogram (XIC) of [366.137 ± 0.0005] Da corresponding to the signature glycan peak in the trypic digest of CAP256-VRC26.25 samples: a distribution of the light chain (Fv) glycopeptides in the control material (top trace); only the heavy chain (Fc) glycopeptides are observed in the CAP256-VRC26.25-N26LCS material (bottom trace). XIC peak intensities have been normalized](/cms/asset/57bf1469-a8c3-47df-aa6a-1ad7055f5107/kmab_a_1836719_f0003_oc.jpg)
Figure 4. Neutralization, thermostability, pharmacokinetics, and heterogeneity of PGT121-S60NHCQ-N68HCT-N105HCK. (a) Neutralization IC50 (µg/ml) of PGT121 wild type and PGT121-S60HCN-N68HCT-N105HCK assessed with nine HIV-1 strains. (b) Pharmacokinetic profile for PGT121 and PGT121-S60HCN-N68HCT-N105HCK in humanized FcRn mice. Dash line denotes limit of detection. (c) Extracted ion chromatogram (XIC) of [366.137 ± 0.0005] Da corresponding to the signature glycan peak in the combined [trypsin + chymotrypsin] digests of PGT121 samples: a distribution of the light chain (Fv) glycopeptides in the control material (top trace); only the heavy chain (Fc) glycopeptides are observed in the PGT121-S60HCN-N68HCT-N105HCK material (bottom trace). XIC peak intensities have been normalized
![Figure 4. Neutralization, thermostability, pharmacokinetics, and heterogeneity of PGT121-S60NHCQ-N68HCT-N105HCK. (a) Neutralization IC50 (µg/ml) of PGT121 wild type and PGT121-S60HCN-N68HCT-N105HCK assessed with nine HIV-1 strains. (b) Pharmacokinetic profile for PGT121 and PGT121-S60HCN-N68HCT-N105HCK in humanized FcRn mice. Dash line denotes limit of detection. (c) Extracted ion chromatogram (XIC) of [366.137 ± 0.0005] Da corresponding to the signature glycan peak in the combined [trypsin + chymotrypsin] digests of PGT121 samples: a distribution of the light chain (Fv) glycopeptides in the control material (top trace); only the heavy chain (Fc) glycopeptides are observed in the PGT121-S60HCN-N68HCT-N105HCK material (bottom trace). XIC peak intensities have been normalized](/cms/asset/c8c05621-30c9-468e-9d74-fb01d6f3448c/kmab_a_1836719_f0004_oc.jpg)
Table 1. Analysis of the presence of somatically acquired N-linked glycosylation sequon for VRC01-like antibody light chains
Figure 5. Design of Fv glycan-removed antibodies without enhanced polyreactivity. (a) Schematic of overall approach. (b) Design strategy
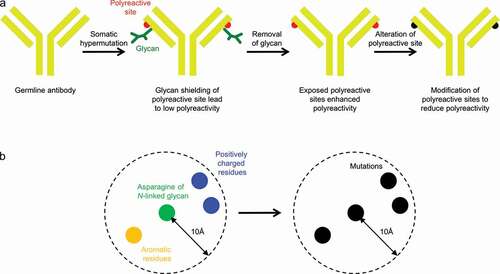
Figure 6. Design of light chain glycan-removed N6 variant. (a) To reduce heightened polyreactivity from removal of light chain glycan 72, mutations were designed to replace aromatic or positively charged residues (shown in yellow) proximal to light chain residue 72 (shown in green). (b) HEp-2 cell staining (antibody concentration: 25 µg/ml) for N6 variants. N6-N72LCQ-R18LCD has similar reactivity as N6 wild type. VRC01LS, 4E10, and VRC07-G54W were used as controls and were assigned to a value between 0 to 3. (c) Anticardiolipin ELISA for N6 variants. N6-N72LCQ-R18LCD showed reduced reactivity as compared to other variants. VRC01LS and VRC07-G54W were used as positive and negative controls, respectively
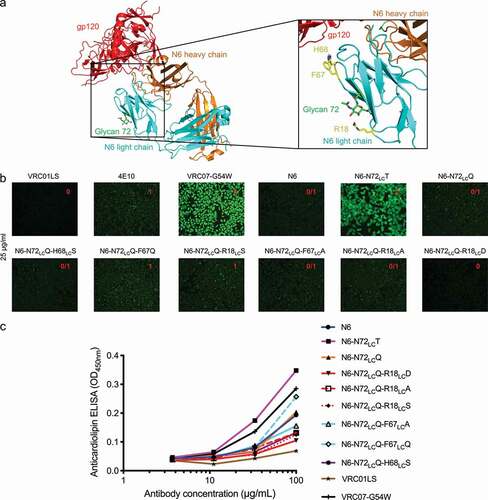
Figure 7. Neutralization, thermostability, pharmacokinetics and heterogeneity of N6-N72LCQ-R18LCD. (a) Neutralization of 208 HIV-1 isolates by N6 wild type and N6-N72LCQ-R18LCD. (b) Differential scanning calorimetry for N6 variants. Removal of glycan did not affect the thermostability of N6. (c) Pharmacokinetic profile for N6LS and N6LS-N72LCQ-R18LCD in humanized FcRn mice. Dash line denotes limit of detection. (d Extracted ion chromatogram (XIC) of [366.137 ± 0.0005] Da corresponding to the signature glycan peak in the combined [trypsin + LysC] digests of N6LS samples: a distribution of the light chain (Fv) glycopeptides in the control material (top trace); only the heavy chain (Fc) glycopeptides are observed in the N6-N72LCQ-R18LCD-LS material (bottom trace). The XIC peak intensities are normalized
![Figure 7. Neutralization, thermostability, pharmacokinetics and heterogeneity of N6-N72LCQ-R18LCD. (a) Neutralization of 208 HIV-1 isolates by N6 wild type and N6-N72LCQ-R18LCD. (b) Differential scanning calorimetry for N6 variants. Removal of glycan did not affect the thermostability of N6. (c) Pharmacokinetic profile for N6LS and N6LS-N72LCQ-R18LCD in humanized FcRn mice. Dash line denotes limit of detection. (d Extracted ion chromatogram (XIC) of [366.137 ± 0.0005] Da corresponding to the signature glycan peak in the combined [trypsin + LysC] digests of N6LS samples: a distribution of the light chain (Fv) glycopeptides in the control material (top trace); only the heavy chain (Fc) glycopeptides are observed in the N6-N72LCQ-R18LCD-LS material (bottom trace). The XIC peak intensities are normalized](/cms/asset/cc925d72-72ad-48f4-a49a-41351d26ce16/kmab_a_1836719_f0007_oc.jpg)
Figure 8. Design of light chain glycan-removed VRC07-523LS variant. (a) To reduce heightened polyreactivity from removal of light chain glycan 72, mutations were designed to replace aromatic or positively charged residues (shown in yellow) proximal to light chain residue 72 (shown in green). (b) HEp-2 cell staining (antibody concentration: 25 µg/ml) for VRC07-523LS variant variants. VRC01LS, 4E10, and VRC07-G54W were used as controls and were assigned to a value between 0 to 3. (c) Anticardiolipin ELISA for VRC07-523LS variants. VRC01LS and VRC07-G54W was used as positive and negative controls, respectively
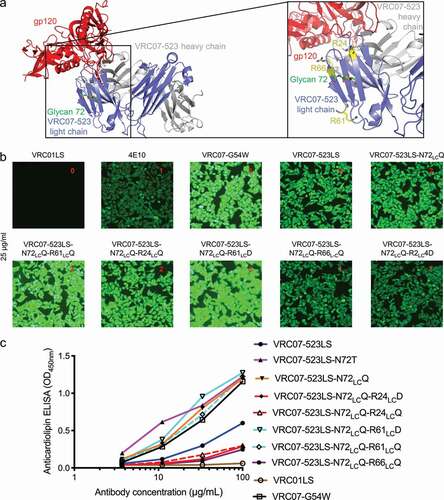
Figure 9. Neutralization, thermostability, pharmacokinetics, and heterogeneity of VRC07-523LS. (a) Neutralization of 10 HIV-1 isolates for VRC07-523LS wild type and VRC07-523LS-N72LCQ-R24LCD. (b) Differential scanning calorimetry for VRC07-523LS variants. Removal of glycan did not affect the thermostability of VRC07-523LS. (c) Pharmacokinetic profile for VRC07-523LS and VRC07-523LS-N72LCQ-R24LCD in humanized FcRn mice. Dash line denotes limit of detection. (d) Extracted ion chromatogram (XIC) of [366.137 ± 0.0005] Da corresponding to the signature glycan peak in the combined [trypsin + LysC] digests of VRC07-523LS samples: a distribution of the light chain (Fv) glycopeptides in the control material (top trace); only the heavy chain (Fc) glycopeptides are observed in the VRC07-523LS-N72LCQ-R24LCD material (bottom trace). XIC peak intensities have been normalized
![Figure 9. Neutralization, thermostability, pharmacokinetics, and heterogeneity of VRC07-523LS. (a) Neutralization of 10 HIV-1 isolates for VRC07-523LS wild type and VRC07-523LS-N72LCQ-R24LCD. (b) Differential scanning calorimetry for VRC07-523LS variants. Removal of glycan did not affect the thermostability of VRC07-523LS. (c) Pharmacokinetic profile for VRC07-523LS and VRC07-523LS-N72LCQ-R24LCD in humanized FcRn mice. Dash line denotes limit of detection. (d) Extracted ion chromatogram (XIC) of [366.137 ± 0.0005] Da corresponding to the signature glycan peak in the combined [trypsin + LysC] digests of VRC07-523LS samples: a distribution of the light chain (Fv) glycopeptides in the control material (top trace); only the heavy chain (Fc) glycopeptides are observed in the VRC07-523LS-N72LCQ-R24LCD material (bottom trace). XIC peak intensities have been normalized](/cms/asset/58f000a7-400d-462a-8cef-bcdb2835d631/kmab_a_1836719_f0009_oc.jpg)
