Figures & data
Figure 1. Therapeutic efficacy of bnAb PGDM1400 in HIS mice. (a) Neutralization sensitivity of BG505, REJO, MJ4 and AMC008 virus to bnAb PGDM1400 in vitro. IC50 and IC90 are indicated by the dotted lines and shown in the table in μg/ml. (b) Viral loads (RNA copies/ml) over time in the HIS mice infected with one of the four viruses. Each line represents one mouse and symbols reflect viral load measurements. The 4 weeks of PGDM1400 administration (20 mg/kg every 7 days) are shaded in gray and arrows for each infection of PGDM1400. (c) The PGDM1400 serum concentration (μg/ml) in each animal in gray with symbols reflecting measurements and the average per group indicated in color. (d) Changes in log10 viral load from start of antibody administration are indicated with each line representing one mouse and the average per group indicated in color
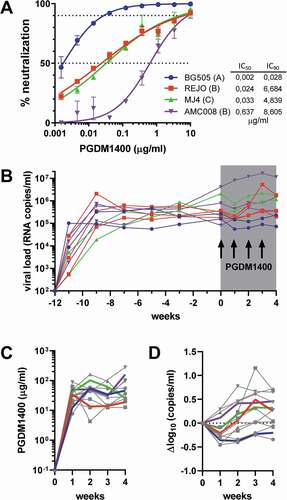
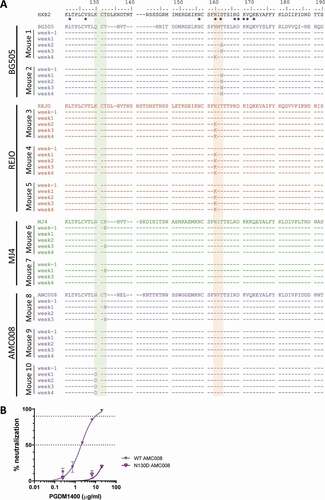
Figure 2. Viral escape over time during PGDM1400 treatment. (a) Amino acid sequences of the HIV-1 envelope glycoprotein (amino acid 75–250) obtained from serum RNA per mouse per time point before and during PGDM1400 treatment. The original virus sequence is indicated above those found in the mice infected with that particular virus. The colors are similar to the colors used in as indicated in Figure 1A. The HXB2 sequence on which numbering is based, is included as reference. Known contact residues for PGDM1400Citation19 are indicated with *. The glycosylation site at position 130 is shaded in green and the one at position 160 in orange. Identical amino acids are indicated by – and gaps by ~. (b) Neutralization sensitivity of AMC008 virus and AMC008 N130D virus to bnAb PGDM1400. IC50 and IC90 are indicated by the dotted lines