Figures & data
Figure 1. Deselection approaches do not prevent positive charge enrichment
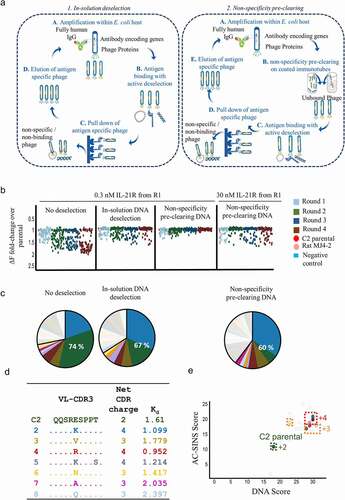
Figure 2. NGS of R3 selection outputs
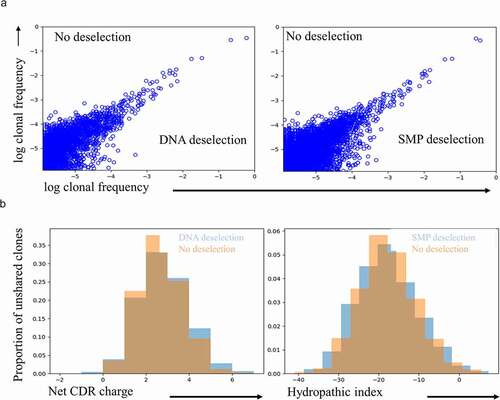
Table 1. Frequency of dominant clones within NGS dataset
Figure 3. Structurally guided interrogation of NGS datasets
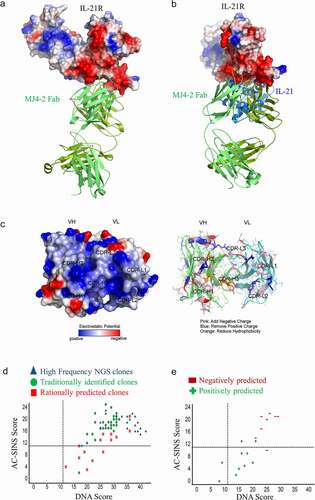
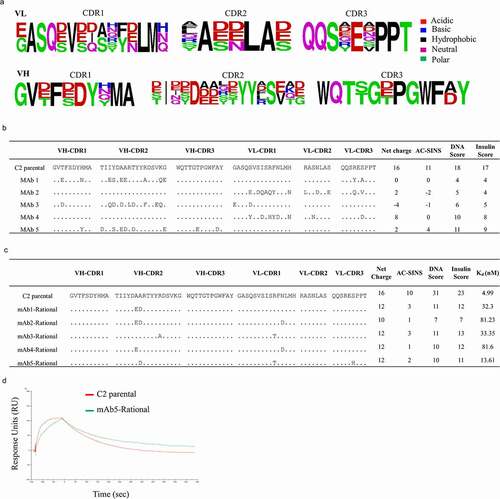
Figure 4. Design, selection and screening of a computationally designed rational library