Figures & data
Figure 1. Schematic workflow of hydrogen/deuterium exchange coupled to mass spectrometry (HDX-MS); binding between the antibody (garadacimab) and antigen (β-FXIIa); and confirmation of binding stoichiometry. Garadacimab, β-FXIIa, and the bound complex were individually incubated in deuterated buffer for different exposure times. The differentially deuterated materials were quenched using a low-pH denaturing and reducing cocktail at low temperature. The quenched mixtures were subsequently digested by pepsin. The reduced and digested peptides from varying exposure times were analyzed using liquid chromatography–mass spectrometry (LCMS) and the mass isotopic data were processed to determine the mass shifts indicative of differential deuterium incorporation (or uptake). Formation of the complex and confirmation of complex integrity were guided by surface plasmon resonance binding kinetics and stoichiometry analysis by negative staining electron microscopy. Data interpretation was performed by mapping the exposure of peptide to solvent against the crystal structure of β-FXIIa and complementarity determining region paratope regions according to Kabat numbering.
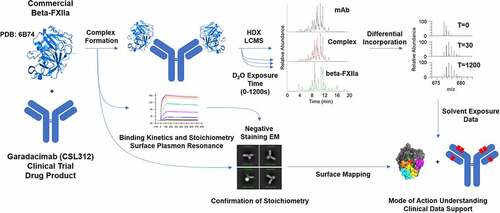
Figure 2. Negative stain electron microscopy images used to visually confirm the stoichiometry of the garadacimab–β-FXIIa complex. (a) The nine panels show the antigen–antibody complex at different angular views, demonstrating the flexibility of the hinge-stabilized IgG4. The extracted pixel box size shows 84 pixels with a pixel size of 4.48 Å. (b) Real space (central) slices of the 3D reconstruction from the three orthogonal viewing directions. The color gradient shows the relative density of the particle image and the axis scale is in pixels with a pixel size of 4.48 Å. (c) Binding of β-FXIIa to captured garadacimab was determined by surface plasmon resonance. Sensorgrams (colored) and fitted curves (black) for each concentration (10 nM–0.02 nM) are shown. Kinetic rate constants and affinity of the interaction are shown in the inset.
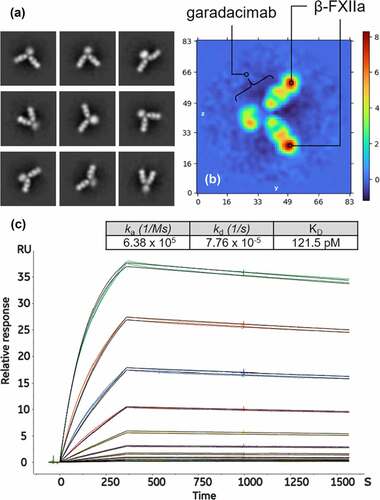
Figure 3. Differential hydrogen/deuterium (H/D) exchange (average relative deuterium) in garadacimab and β-FXIIa at various exposure times (30–1200 seconds). Specific paratopes on (a) garadacimab light chain complementarity determining region (CDR) and (b) garadacimab heavy chain CDR and the expected non-paratopic Fc control region. (c) β-FXIIa with six epitopic regions highlighted (red: histidine residue of the catalytic triad; blue: N-terminus of β-FXIIa; Orange: 99-loop; magenta: autolysis 140-loop; cyan: activation 180-loop; and yellow: 220-loop).
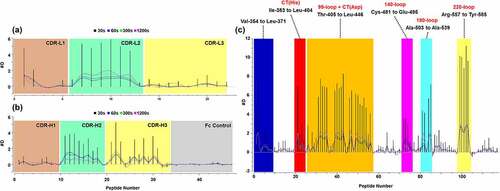
Figure 4. (a) Peptic peptide fragment ion coverage of the heavy chain variable region and the Fc constant heavy region 2 (CH2) constant region. (b–i) Kinetics of the hydrogen/deuterium (H/D) exchange showing the percent of deuterium exchanged as a function of time for selected peptides. Lines are drawn as a guide to the eye (black: free garadacimab; green: complexed garadacimab). Asterisks denote significance of change at the 95% confidence interval. CDR, complementarity determining region.
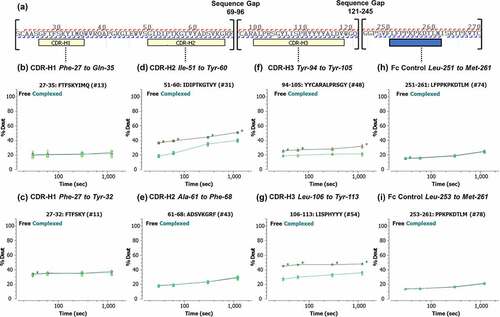
Figure 5. (a) Mapping of hydrogen/deuterium (H/D) exchange on regions of β-FXIIa (structure based on X-ray crystallography).Citation24 Separate epitopic regions on β-FXIIa based on HDX-MS data are highlighted (red: catalytic triad; blue: N-terminus of β-FXIIa; green: 60-loop, Orange: 99-loop; magenta: autolysis 140-loop; cyan: activation 180-loop; and yellow: 220-loop). Additional designation of the S1 pocket is shown to demonstrate potential blocking of substrate entry. (b–g) Corresponding kinetics of H/D exchange in β-FXIIa via selected peptides covering denoted epitopic regions. (h) Solvent-exposed 110-loop (as negative control), showing no significant difference in deuterium uptake. Lines are drawn as a guide to the eye (black: free β-FXIIa; green: complexed β-FXIIa). Asterisks denote significance of change at the 95% confidence interval.
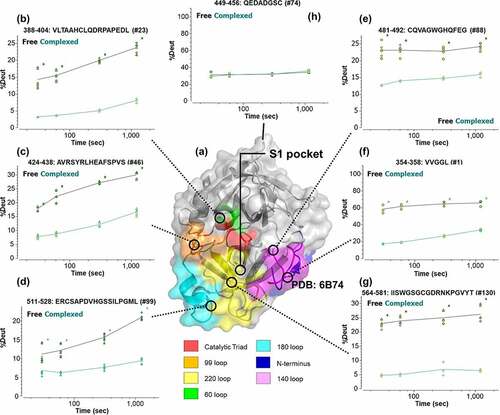
Figure 6. Sequence alignment and mapping of the hydrogen/deuterium (H/D) exchange data and volume area dihedral angle reporter (VADAR) analysis (with side-accessibility surface area [side-ASA] of ≥50%) on regions of β-FXIIa. VADAR analyses were computed using X-ray crystallography coordinates from the protein data bank.Citation24 Red boxes denote regions identified by HDX-MS data while bolded text denotes surface-exposed residues (from VADAR analysis). Various surface loops and termini with notable reduction in H/D uptake (N-terminus, 60-loop, 99-loop, 140-loop, 180-loop, and 220-loop) are shown. Asterisks denote the position of the catalytic triad His-Asp-Ser on β-FXIIa.
![Figure 6. Sequence alignment and mapping of the hydrogen/deuterium (H/D) exchange data and volume area dihedral angle reporter (VADAR) analysis (with side-accessibility surface area [side-ASA] of ≥50%) on regions of β-FXIIa. VADAR analyses were computed using X-ray crystallography coordinates from the protein data bank.Citation24 Red boxes denote regions identified by HDX-MS data while bolded text denotes surface-exposed residues (from VADAR analysis). Various surface loops and termini with notable reduction in H/D uptake (N-terminus, 60-loop, 99-loop, 140-loop, 180-loop, and 220-loop) are shown. Asterisks denote the position of the catalytic triad His-Asp-Ser on β-FXIIa.](/cms/asset/1e660f8c-058e-4010-a8f9-38a7aaab9534/kmab_a_2163459_f0006_oc.jpg)
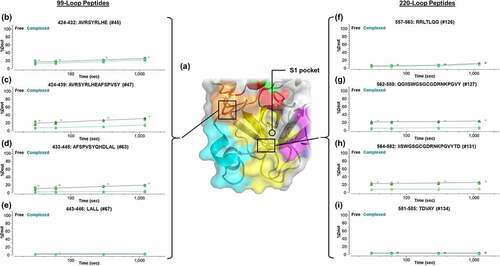
Figure 7. Localization of the epitope region specific to the entrance to the S1 substrate pocket via the 99-loop and 220-loop through the utilization of overlapping peptic peptides. (a) Correlation with the X-ray crystallography-based structure of β-FXIIa.Citation24 Kinetics of deuteration are shown in (b–e) for the 99-loop region and (f–i) for the 220-loop. Lines are drawn as a guide to the eye (black: free β-FXIIa; green: complexed β-FXIIa). Asterisks denote significance of change at the 95% confidence interval.