Figures & data
Figure 1. Approaches to discover antibody candidates with fewer physicochemical liabilities. a Rationally designed antibody libraries can be created by selecting HCDR1-2 and LCDR1-3 sequences without known sequence-based liabilities, screening the selected sequences as single-CDR libraries for expression on yeast and combining the sequences with well-behaved human scFv scaffolds and diverse HCDR3 sequences obtained from human donors. b VHH phage libraries can be depleted of aggregation-prone sequences before biopanning to increase the probability of selecting VHH with resistance to aggregation. c Integration of antibody genes into a single locus allows transcriptional normalization and comparison of antibody variants based on their expression level on the surface of mammalian cells. High display level of the antibodies is correlated with favorable biophysical properties.
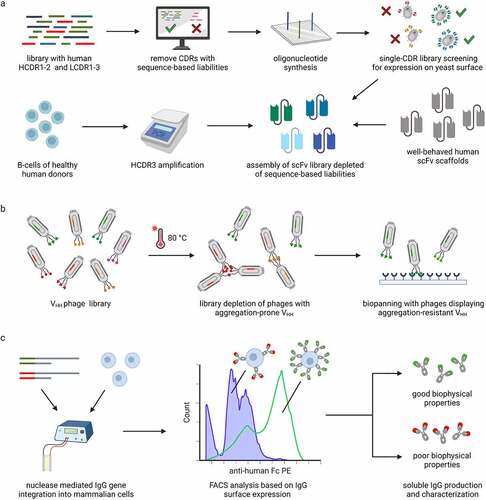
Figure 2. Approaches to select mutations that optimize antibody properties. a Schematic overview of phage assisted continuous evolution. Host bacteria contain accessory plasmid with the pIII phage gene needed for infectivity and a mutagenesis plasmid. The host bacteria flow constantly through a vessel (“lagoon”) containing selection phages that contain the gene of interest (GOI). The production of the pIII is dependent on properties of the GOI. Only selection phages that encode functional variants of the GOI produce pIII and infectious progeny that can reinfect new host bacteria to remain in the “lagoon”. Phages with non-functional variants cannot infect new host bacteria before being washed away from the flow. b In the tripartite β-lactamase enzyme assay, scFv sequences are inserted between two domains of β-lactamase. If the scFv are resistant to aggregation in the bacterial periplasm, the split β-lactamase can fold into a functional form and the bacteria gain resistance to ampicillin. Selection of different variants can be made by assessing the growth of bacterial clones on agar plates with different ampicillin concentrations. c Machine learning model trained on a library with >107 emibetuzumab Fab variants sorted for binding affinity and specificity can predict mutations that co-optimize antibody affinity and specificity.
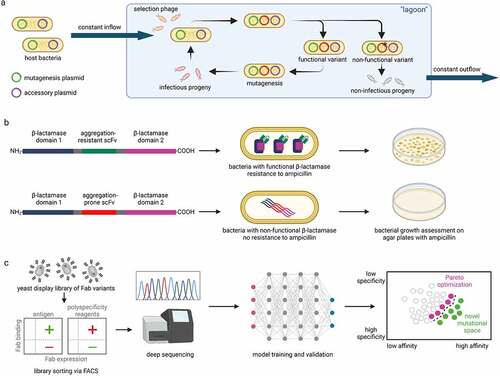
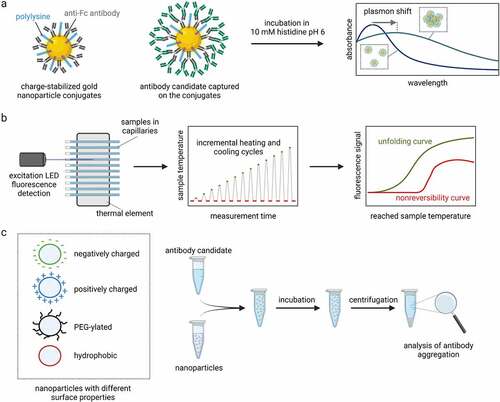
Figure 3. Methods to select antibody candidates with desired physicochemical properties. a Charge-stabilized self-interaction nanoparticle spectroscopy (CS-SINS) can assess the self-association of antibody candidates in common pharmaceutical buffers (10 mM histidine pH 6) at ultra-dilute antibody concentrations (0.01 mg/mL). The CS-SINS score is calculated from the plasmon shift due to self-association of the nanoparticle conjugates and can predict solution properties such as viscosity and opalescence at high antibody concentrations. b Modulated scanning fluorimetry (MSF) employs incremental heating and cooling cycles to simultaneously probe the thermal unfolding and unfolding reversibility of dozens of antibody candidates by consuming only 10 µL per condition. c The nanoparticle-surface mediated stress assay uses nanoparticles with different surface properties. The nanoparticles are mixed and incubated with antibody candidates. The large nanoparticle surface accelerates the surface-induced aggregation of antibodies and allows ranking of candidates based on their interfacial stability.