Figures & data
Figure 1. Design of HIV-1 bispecific antibodies attaching nanobodies or scFvs to the light chain of the super potent V2-apex-directed antibody CAP256-VRC26.25. (a) Structure of the antigen-binding fragment (Fab) of antibody CAP256-VRC26.25 in complex with a prefusion-closed HIV-1 Env trimer showing an unencumbered light chain allowing its linkage to other HIV-1 trimer-binding components. The resultant bispecific antibody enables synergistic binding and enhanced neutralization. (b) Schematic of V1V2 antibody with additional binding component genetically fused to the light chain N terminus. (c) Light chain variant expression constructs. Sequence information of these variants are listed in supplementary figures. Upper construct shows light chain variants linked with nanobody. Bottom construct shows light chain variants linked to single-chain Fv.
Figure 2. Evaluation of antibody variants and selection of variants with improved neutralizing breadth. (a) Screening of designed antibody variants for binding to trimers from CAP256V2LS-resistant virus strains and neutralization on a CAP256V2LS-resistant virus. Each row represent the results of binding in green color or neutralization in red color. Darker colors indicate better binding or neutralization. (b) Neutralization IC50 of CAP256V2LS nanobody variants and scFv variants on small virus panels. (c) Neutralization IC50 of PGDM1400 antibody variants on a 5-virus panel. (d) Neutralization IC80 of selected CAP256V2LS and PGDM1400 antibody variants on a 30-virus panel. Geometric mean IC80 values (µg/ml) are indicated at the bottom of each antibody column.
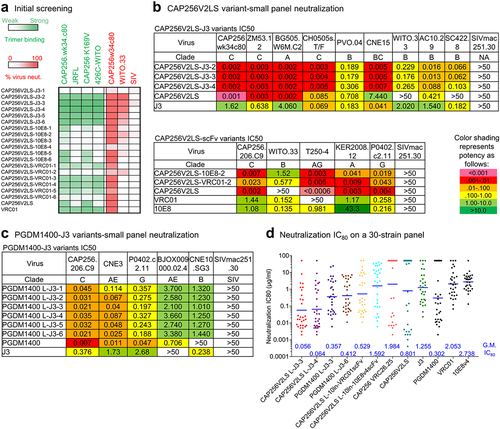
Figure 3. Structure of CAP256V2LS-J3-3 Fab in complex with BG505 DS-SOSIP.664 confirms avidity and stoichiometry. (a) Cryo-EM density is shown highlighting the linker between CAP256V2LS and J3 at 3.2 Å resolution and contour of 0.22 where density for the flexible linker was visible. (b) Cryo-EM density is shown with lower contour to illustrate the signal observed corresponding to unbound CAP256V2LS in Orange (c) (left) Cryo-EM density is shown with higher contour revealing greater detail of higher resolution signal. (right) Corresponding atomic models are shown in cartoon format with glycans shown as spheres. (d) Details of binding for CAP256V2LS and J3 from the bivalent complex structure are overlayed with the structures obtained from individual components. (e) The CDR H3 of CAP256V2LS aligns closely while the main body of the Fab shifts as much as ~16 Å.
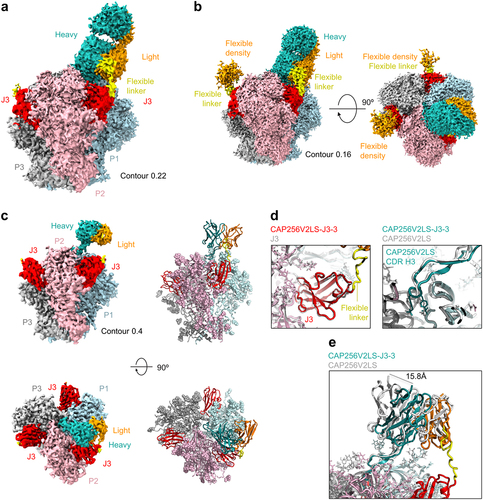
Figure 4. Improved pharmacokinetics of CAP256.J3LS achieved by altering surface charge of J3. (a) In vivo half-life of CAP256- variants assessed in a human FcRn knock-in mouse model. (b) Sequence of J3 with Arg and Lys residues highlighted, and J3 paratope residues are underlined. (c) Accessible surface area of Arg and Lys residues. Residues above the dotted line were altered mutationally to reduce electropositivity. (d) Arg and Lys residues that were selected for mutations were shown in the structure of J3 in complex with gp120 (PDB ID: 7RI1). (e) Neutralization IC80 fold change, heparin chromatography retention volume, and autoreactivity of CAP256.J3LS variants. (f) In vivo half-life of CAP256.J3LS variants.
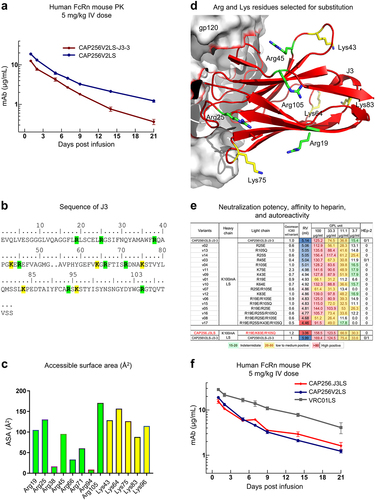
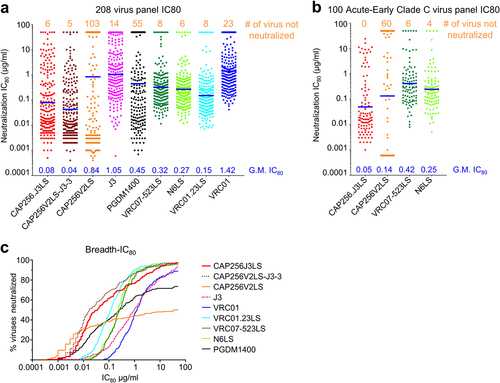
Figure 5. Neutralization breath and potency of CAP256.J3LS antibody. (a) Neutralization IC80 of CAP256.J3LS on a 208 global virus panel in comparison with parental antibodies and select HIV-1 antibodies in clinical development. (b) Neutralization IC80 of CAP256.J3LS on a 100 Acute-Early Clade C virus panel. (c) Breadth-IC80 curves of CAP256.J3LS in comparison with parental antibodies and select HIV-1 antibodies in clinical development.
Supplemental Material
Download Zip (6.6 MB)Data availability statement
Cryo-EM maps and fitted coordinates have been deposited with EMDB entry ID EMD-29209 (https://www.ebi.ac.uk/emdb/search/EMD-29209) and PDB entry ID 8FIS (https://www.rcsb.org/structure/unreleased/8FIS), respectively.
