Figures & data
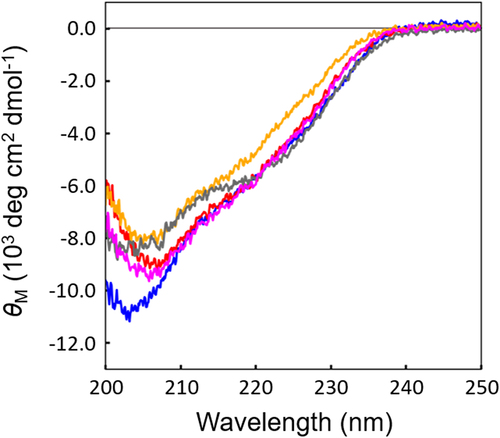
Figure 1. Three-dimensional structure of the entire sequence of 2u2f. The two randomized loops are in red.
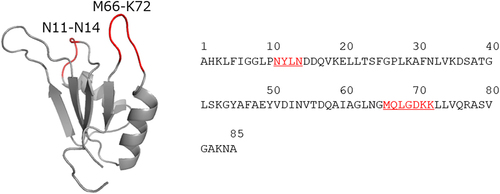
Figure 2. Workflow of biopanning. At each round, 1) target-bound phages were selected, 2) E. coli was infected with selected phages, and 3) phages were amplified in E. coli. Sub-libraries are surrounded by colored ellipses.
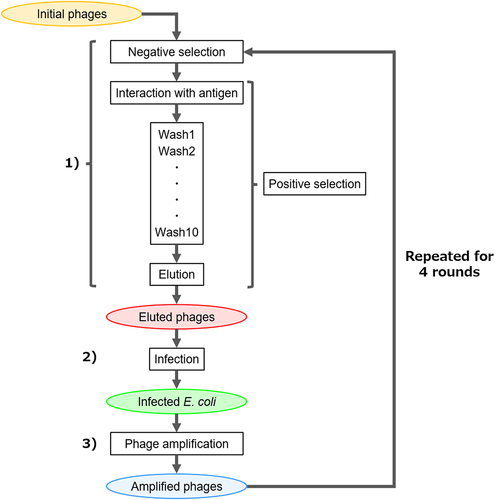
Figure 3. Distribution of unique sequences in each sub-library. The frequency of unique sequences is shown for single reads in gray, 2–10 reads in blue, 11–100 reads in green, 101–200 reads in yellow, 201–1000 reads in brown, and >1000 reads in red.
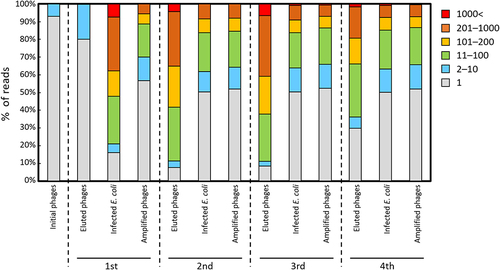
Figure 4. Amino acid frequencies and rank distribution of the sequences predicted by machine learning. (a) Amino acid frequencies of top 10,000 sequences predicted by machine learning, visualized by WebLogo.Citation41 (b) Amino acid frequencies of clustered sequences. (c) Rank distribution of each cluster. Black arrows indicate clusters containing the top 1,000 sequences.
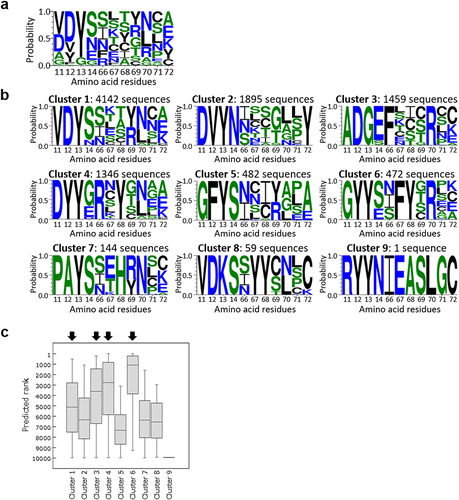
Figure 5. Binding function of wild-type 2u2f and obtained 2u2f variants. (a) Enzyme-linked immunosorbent assay of the candidate 2u2f variants after purification on galectin-3 (Gal), NeutrAvidin (NAV), or blocking buffer (Skim). (b) EC50 values of wild-type 2u2f and four functional variants with affinity to galectin-3. The plots show the absorbance of galectin-3 minus that of NAV. The EC50 values were determined by using Hill equation.
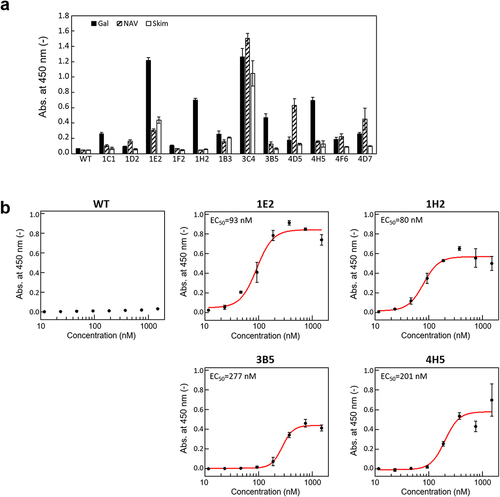
Figure 6. CD spectra of the functional 2u2f variants. Wild-type 2u2f is shown in blue, 1E2 in Orange, 1H2 in red, 3B5 in gray, and 4H5 in magenta.