Figures & data
Figure 1. ZPDGFRb_3 binds PDGFRB without inducing receptor activation or internalization. (a) MG − 63 cell lines are positively stained by an anti-PDGFRB antibody via a secondary FITC (fluorescein isothiocyanate)-conjugated secondary antibody, compared to FITC-conjugated secondary antibody alone. (b) ZPDGFRb_3 binds MG − 63 cells when compared to an Affibody molecule binding an irrelevant target. (c) BUD − 8 cell line is stimulated with PDGF-BB (PDGFRB natural ligand) or ZPDGFRb_3 and cell proliferation is assessed after 48 hours. ZPDGFRb_3 does not stimulate cell proliferation when compared to the natural ligand. (d) BJhTERT cells are treated with either ZPDGFRb_3 or PDGF-BB, at 37°C or on ice to assess PDGFRB activation and internalization-dependent degradation by western blot. Receptor phosphorylation (P-Tyr) is observed in the presence of PDGF-BB but not with ZPDGFRb_3 (performed on ice to prevent receptor internalization). Lower levels of PDGFRB due to receptor internalization-dependent degradation are observed in presence of PDGF-BB but not with ZPDGFRb_3.
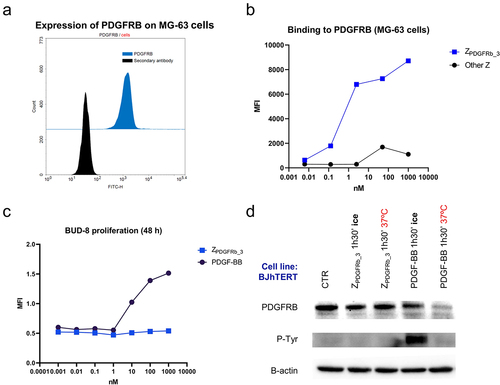
Figure 2. The bispecific PDGFRBxCD40 AffiMab binds both PDGFRB and CD40. (a) the AffiMab is designed from a CD40 agonistic, LALAPG-mutated antibody (unable of binding the FcγR). One ZPDGFRb_3 is linked to the C-terminus of each heavy chain. (b) the binding properties of the AffiMab are tested against CD40 and PDGFRB by SPR. (c) Simultaneous binding of the AffiMab toward PDGFRB and CD40 confirmed by BLI. (d) All the three molecules in (a) bind the CD40 expressing cell line HEK (human embryonic kidney)-Blue CD40L while only the AffiMab binds PDGFRB expressing cells MG − 63, assessed by flow cytometry.
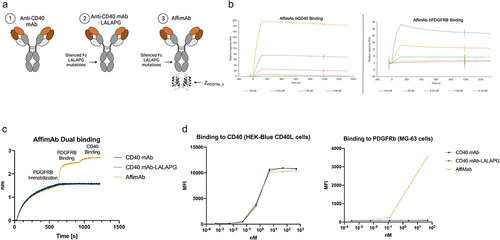
Figure 3. PDGFRB-targeted CD40 activation in HEK-Blue CD40L reporter gene cell assay. (a) Schematic illustration of the assay using HEK-Blue CD40L cells as reporter gene cell assay with either PDGFRB-conjugated, PDGFRB-conjugated beads in different ratios with Chicken IgY and unconjugated beads or mono and co-cultures in presence of the AffiMab. (b) CD40 activation occurs at lower AffiMab concentrations in presence of PDGFRB conjugated beads compared to unconjugated beads. (c) PDGFRB-targeted CD40 activation is dependent on PDGFRB availability/density on the beads. Beads were used with PDGFRB at a ratio of 1/4, 2/4, or 3/4 of the total amount of protein bound to the beads. An anti-chicken IgY biotinylated antibody was used to occupy the beads together with PDGFRB. Beads carrying only PDGFRB or anti-chicken IgY antibody were also included. The CD40 activation signal intensity dependent on PDGFRB availability on the beads. Mean and standard deviation of one representative experiment of at least two independent replicates are displayed on (B-C). (d) MG − 63 Co-culture (f). NIH − 3T3 Co-culture. Mean and standard deviation of two independent replicates shown (D-E).
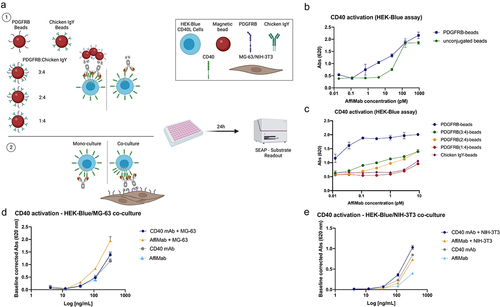
Figure 4. AffiMab mediated, PDGFRB-dependent CD40 activation in moDcs. (a) in a similar fashion as in , the AffiMab is evaluated on moDcs in presence of PDGFRB-conjugated or unconjugated beads. The CD40 mAb-LALAPG is also tested in the presence of PDGFRB-conjugated beads. The same experiment is repeated using BJhTERT cells as source of PDGFRB, performing a mono and co-culture. (b) Only the AffiMab activates moDcs in bead-based set-up specifically in presence of PDGFRB, as measured by upregulation of CD86 in flow cytometry. (c) CD86 is upregulated on moDcs following AffiMab treatment only in presence of PDGFRB expressing cells. Average mean fluorescence intensity (MFI) of one representative experiment of at least two independent replicates is displayed on (B-C).
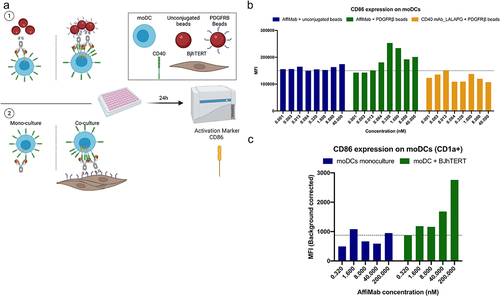
Figure 5. AffiMab mediated, PDGFRB-dependent CD40 activation in B cells. (a) B cells are co-cultured with either PDGFRB+ BJhTERT or PDGFRB− A431 cells in presence of the AffiMab. (b) PDGFRB expression is evaluated on BJhTERT and A431 cells by flow cytometry. The former is confirmed PDGFRB+ as also shown by western blot data in . A431 cells are identified as PDGFRB−. Unstained cells are included as negative control. (c) CD86 fraction of CD20+ B cells is higher at lower AffiMab concentrations when PDGFRB is provided by cells. One representative experiment of at least two independent replicates is plotted with CD86+ cells (on CD20+ B cells) calculated on at least 1 × 104 collected cell events.
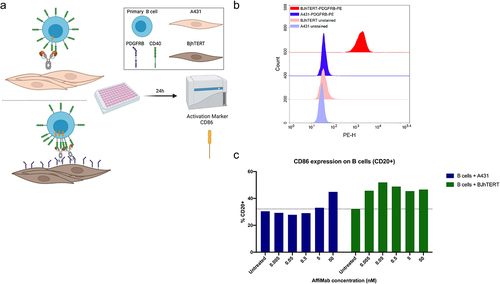
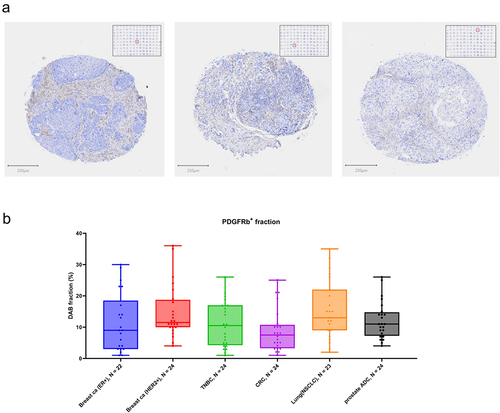
Figure 6. PDGFRB immunohistochemical staining of clinical tumor specimens. Staining settings: PDGFRB-DAB (brown) and Hematoxilin (blue). (a) Representative tumor cores of a tumor TMA, showing different patterns of PDGFRB in the tumor stroma. (b) Quantification of PDGFRB fraction (positive staining area as percentage tumor core area) showing overall representation of PDGFRB staining across different tumor types.