Figures & data
Figure 1. Characterization of Group 1 ILCs within the human colonic lamina propria. (a) Representative flow cytometry gating strategy to identify NK cells and ILC1s in human colonic tissue. (b) Frequencies of NK cells and ILC1s in the colon lamina propria layer as a percent of viable CD45+ cells and (c) as number of cells per gram of colonic mucosa ex vivo. N = 7. (d) Representative flow cytometry depicting the expression of T-bet and EOMES by NK cells and ILC1s. (e) Net geometric mean fluorescent intensity of NK cells and ILC1s expressing the transcription factors T-bet (N = 7) or (f) EOMES (N = 4) ex vivo. (g) Percentages of NK cells and ILC1s expressing the EOMES (N = 4) ex vivo. Bars are mean + S.E.M. Statistical analysis performed was paired t-test. *p < .05, **p < .01.
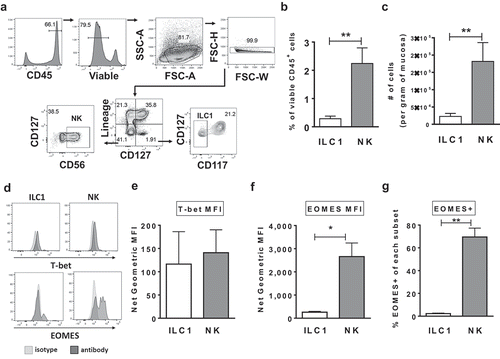
Figure 2. S. typhimurium induces IFNγ and TNFα production by human colonic Group 1 ILCs. (a) Representative flow cytometry demonstrating cytokine staining for IFNγ gated on NK cells or ILC1s after LPMC exposure to S. typhimurium (St) or no bacterial control (-) in vitro. (b) Percentages of IFNγ+ NK cells or ILC1s after LPMC exposure to S. typhimurium or no bacterial control. N = 10. (c) Representative flow cytometry demonstrating cytokine staining for TNFα gated on NK cells or ILC1s after LPMC exposure to S. typhimurium or no bacterial control in vitro. (d) Percentages of TNFα+ NK cells or ILC1s after LPMC exposure to S. typhimurium or no bacterial control. N = 7. Bars are mean + S.E.M. Statistical analysis performed was one-way ANOVA comparing the mean of each column. *p < .05, **p < .01, ***p < .001, n.s. = not significant.
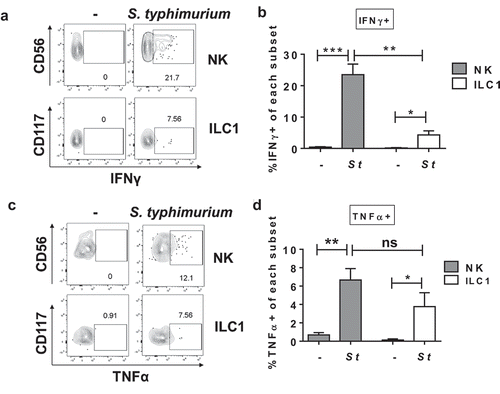
Figure 3. The cytokines IL-12p70, IL-18 and IL-1β contribute to the induction of IFNγ by Group 1 ILCs in response to S. typhimurium. (a) Amount (pg/mL) of IL-12p70, IL-18, or IL-1β secreted into the supernatant after LPMC exposure to S. typhimurium (St) or no bacterial control (-) in vitro. N = 6. (b) Percentages of IFNγ+ NK cells or (c) ILC1s after LPMC exposure to no bacteria control or S. typhimurium in the presence of 10ug/mL blocking antibodies targeting IL-12p70, IL-18, IL-1β or the antibody isotype control IgG. N = 3. Bars are mean + S.E.M. Statistical analysis performed was (a) paired t-test or (b) one-way ANOVA comparing the mean of each column with the mean of the control column S. typhimurium + IgG isotype control. *p < .05, **p < .01.
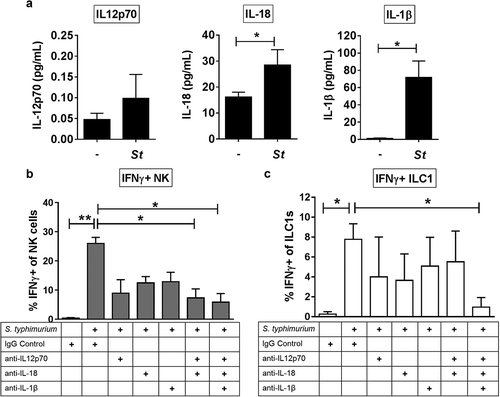
Figure 4. Differential IFNγ induction by Group 1 ILCs in response to a panel of enteric commensal bacteria. (a) Representative flow cytometry demonstrating cytokine staining for IFNγ gated on NK cells or ILC1s after LPMC exposure to Gram-negative A. junii or P. stercorea in vitro. (b) Percentages of IFNγ+ NK cells or ILC1s after LPMC exposure to no bacterial control (-), A. junii (Aj) or P. stercorea (Ps) N = 7. (c) Representative flow cytometry demonstrating cytokine staining for IFNγ gated on NK cells or ILC1s after LPMC exposure to Gram-positive B. infantis or R. bromii in vitro. (d) Percentages of IFNγ+ NK cells or ILC1s after LPMC exposure to no bacterial control or B. infantis (Bi) or R. bromii (Rb). N = 6. Bars are mean + S.E.M. Statistical analysis performed was one-way ANOVA comparing the mean of each column. *p < .05, n.s. = not significant.
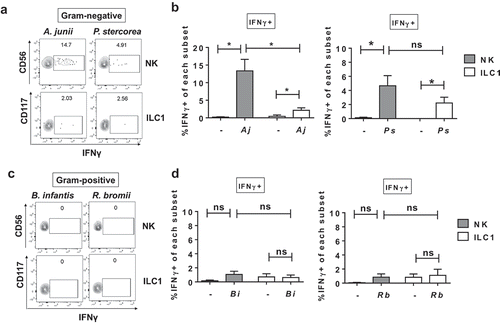
Figure 5. IFNγ production was not mediated by Group 1 ILC direct recognition of Gram-negative bacteria. (a) Percentages of LPMCs stained ex vivo for TLR2, TLR4, or TLR5 expression and gated on NK cells or ILC1s, or Lineage-positive cells that are larger on Forward Scatter-area vs Side Scatter-area flow plots.N = 6. (b) Quantification of IFNγ (pg/mL) in the supernatant of purified NK cells or (c) ILC1s exposed to recombinant IL-12p70 and IL-18 (50ng/mL), A. junii (Aj) or R. bromii (Rb) in vitro. N = 3. (d) Normalization of secreted IFNγ (pg/mL) per the amount of plated NK or ILC1s (cells/mL) to determine IFNγ (pg/cell). N = 3. Bars are mean + S.E.M. Statistical analysis performed was paired t-test or one-way ANOVA comparing the mean of each column. **p < .01, ***p < .001, n.s. = not significant.
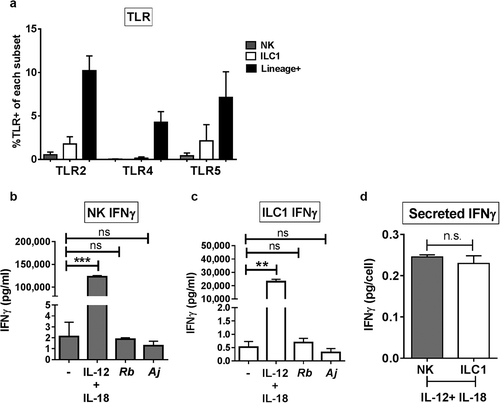
Figure 6. Myeloid dendritic cells producing cytokines in response to Gram-negative commensal bacteria promote IFNγ production by Group 1 ILCs. (a) Percentages of IL-12p70+, IL-18+, or IL-1β+ mDCs or macrophages after LPMC exposure to no bacteria control (-), A. junii (Aj) or R. bromii (Rb). N = 4. (b) Percentages of IFNγ+ NK cells or ILC1s after LPMC exposure to A. junii with no depletion control (total LPMCs) or mDC depletion. N = 3. Bars are mean + S.E.M. Statistical analysis performed was one-way ANOVA comparing the mean of each column with the mean of the control column (no bacteria). *p < .05, #p = .0637.
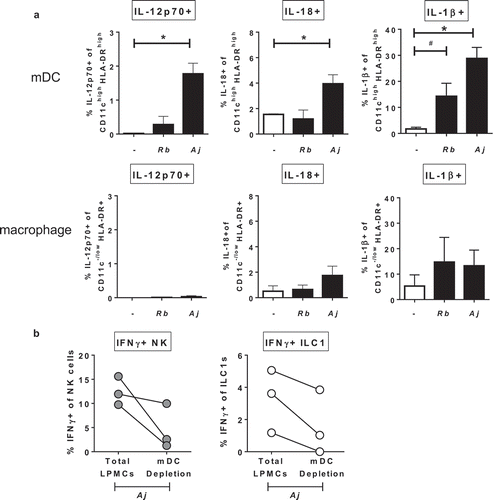
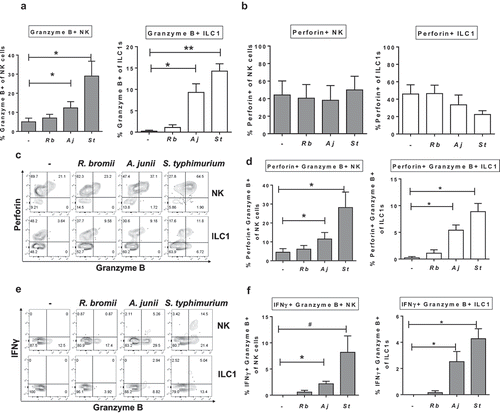
Figure 7. Gram-negative bacteria induce Granzyme B-expressing Group 1 ILCs. (a) Percentages of Granzyme B+ or (b) Perforin+ NK cells or ILC1s after LPMC exposure to no bacteria control (-), R. bromii (Rb), A. junii (Aj) or S. typhimurium (St) in vitro. N = 4. (c) Representative flow cytometry demonstrating co-staining for Granzyme B and Perforin gated on NK cells or ILC1s after LPMC exposure to no bacteria control, R. bromii, A. junii, or S. typhimurium in vitro and (d) Percentages of NK cells or ILC1s expressing both Perforin and Granzyme B. N = 4. (e) Representative flow cytometry demonstrating co-staining for Granzyme B and IFNγ gated on NK cells or ILC1s after LPMC exposure to no bacteria control, R. bromii, A. junii, or S. typhimurium in vitro and (f) Percentages of NK cells or ILC1s expressing both IFNγ and Granzyme B. N = 4. Bars are mean + S.E.M. Statistical analysis performed was one-way ANOVA comparing the mean of each column with the mean of the control column (no bacteria). *p < .05, **p < .01, #p = .0803.