Figures & data
Table 1. Colorectal carcinogenic pathways and main genetic alterations.
Table 2. Hallmarks of cancer and examples of genes silenced by aberrant methylation.
Figure 2. Mechanisms of DNA demethylation. (a) Passive demethylation. This process occurs during replication, wherein one or more limiting factors (i.e., compromised DNMT function, absence of SAM) prevents methylation maintenance and results in the subsequent loss of 5mC residues. (b) Active demethylation. The figure shows TET enzymes (TET1, TET2, or TET3) (teal) catalyzing stepwise oxidation of 5mC, which is first converted into 5-hydoxymethylcytosine (5hmC), further oxidized into 5-formylcytosine (5fC), and finally converted into 5-carbocylcytosine (5caC). 5fC and 5caC intermediates can be recognized and removed by thymine DNA glycosylase (TDG) (violet). They are then replaced with an unmethylated cytosine nucleotide to complete the base excision repair (BER) process (figure from ref).Citation57
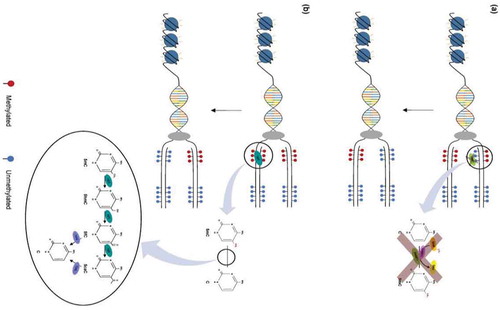
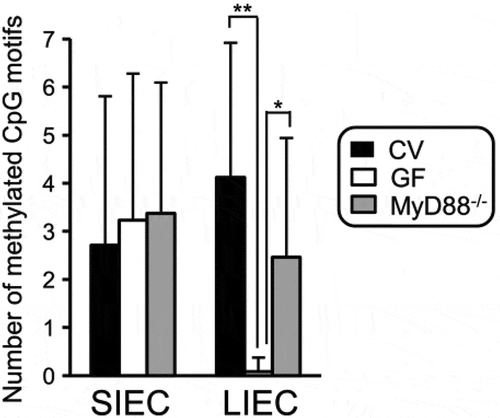
Figure 3. IEC: intestinal epithelial cell; SIEC: small; IECL: IEC large intestine; CV: conventional; GF: germ-free; IEC: intestinal epithelial cell; SIEC:small; LIEC: large intestine; CV:conventional; GF: germ-free; from Takashi et al Ref. Citation94.