Figures & data
Figure 1. A gene encoding 12β-HSDH completes the gut microbial epi-bile acid pathway. Cholic acid (CA) is converted to the oxo-intermediate, 7-oxodeoxycholic acid (7-oxoDCA), and further to ursoCA through the urso-bile acid pathway catalyzed by NAD(P)-dependent 7α- and 7β-HSDH. The secondary bile acid deoxycholic acid (DCA) is formed through the multi-step 7α-dehydroxylation of CA. DCA is biotransformed to 3-oxoDCA by 3α-HSDH and to isoDCA by 3β-HSDH in the iso-bile acid pathway. DCA can be converted to 12-oxolithocholic acid (12-oxoLCA) by 12α-HSDH and from 12-oxoLCA to epiDCA by 12β-HSDH. Examples of bacteria expressing each HSDH are shown below the reaction followed by corresponding gene annotations. Prior to this study, a gene encoding 12β-HSDH had not been identified
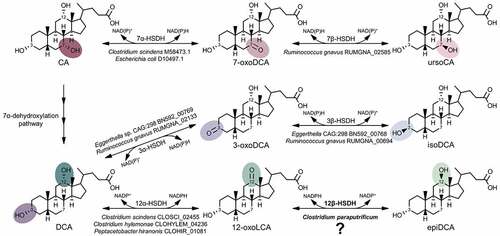
Figure 2. Clostridium paraputrificum ATCC 25780 expresses 12β-HSDH while C. scindens ATCC 35704 expresses 12α-HSDH by whole-cell LC-MS. (a) Representative negative ion mode LC-MS chromatograms in single ion monitoring mode overlaid with linked vertical axes of C. paraputrificum reaction products from 50 μM substrate compared to deoxycholic acid (DCA), 12-oxolithocholic acid (12-oxoLCA) and epiDCA standards. (b) As a control, representative negative ion mode LC-MS chromatograms in single ion monitoring mode overlaid with linked vertical axes of C. scindens products from 50 μM substrate was used to demonstrate 12α-HSDH activity. Standards are shown in (a) and (b) for ease of comparison to products. Formula weight for DCA is 392.57 atomic mass units (amu), 12-oxoLCA is 390.56 amu, epiDCA is 392.57 amu
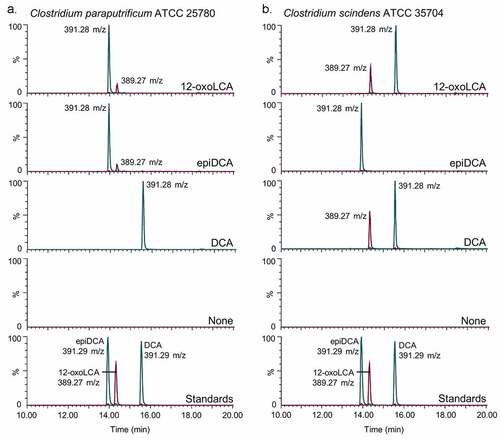
Figure 3. Identification of a gene encoding bile acid 12β-HSDH. (a) SDS-PAGE of candidate Clostridium paraputrificum 12β-HSDH proteins that were heterologously expressed in E. coli and purified with TALON® metal affinity resin. Lanes are as follows: S, molecular weight protein standard; 077, WP_027099077.1; 355, WP_027098355.1; 937, WP_027097937.1; 604, WP_027098604.1; 909, WP_027096909.1; 631, WP_027099631.1. (b) Representative negative ion mode LC-MS chromatograms in single ion monitoring mode overlaid with linked vertical axes of WP_027099077.1 reaction products compared to deoxycholic acid (DCA), 12-oxolithocholic acid (12-oxoLCA) and epiDCA standards. Standards were run on a separate day and show a slight offset in elution time. Reactions consisted of 10 nM WP_027099077.1 with 50 μM (or no) substrate, 150 μM pyridine nucleotide in 50 mM sodium phosphate, 150 mM sodium chloride buffer at pH 7.0. Formula weight for DCA is 392.57 atomic mass units (amu), 12-oxoLCA is 390.56 amu, epiDCA is 392.57 amu
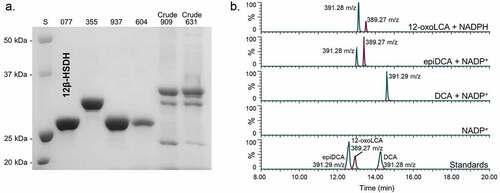
Figure 4. Biochemical characterization of recombinant C. paraputrificum 12β-HSDH. (a) Native molecular size analysis of 10 mg/mL purified 12β-HSDH via size-exclusion chromatography. (b) Effect of pH on 12β-HSDH activity. The reaction in the reductive direction (blue) consisted of 12-oxoLCA as substrate with NADPH as cofactor. The oxidative reaction (red) was epiDCA with NADP+. See Materials and Methods for buffer compositions
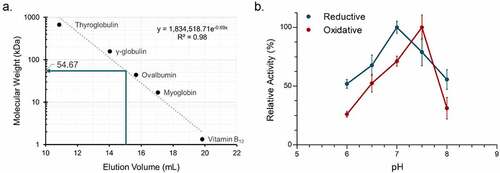
Table 1. Steady-state kinetic parameters of purified recombinant 12β-HSDH
Table 2. Substrate and pyridine nucleotide specificity of purified recombinant C. paraputrificum 12β-HSDH, Eisenbergiella WP_118677302.1, Olsenella WP_120179297.1, and Novosphingobium WP_007678535.1
Figure 5. Maximum-likelihood tree based on a subset of the taxa present in the full phylogenetic analysis of 12β-HSDH and SDS-PAGE of proteins explored further. Sequences selected for this analysis were those nearest to the C. paraputrificum 12β-HSDH (highlighted), plus an outgroup. For the full tree with about 5,000 sequences, see Figure S2. Taxonomic affiliations are indicated by branch colors as specified in the legend. Bolded sequences were chosen for further study. Asterisks indicate novel C-12 epimerizing organisms. (Inset) SDS-PAGE of purified recombinant Eisenbergiella WP_118677302.1, Olsenella WP_120179297.1, and Novosphingobium WP_007678535.1 heterologously expressed in E. coli and purified with TALON® metal affinity resin. S, molecular weight protein standard
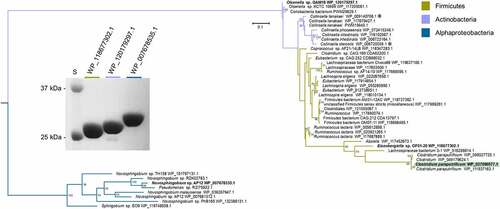
Figure 6. 12β-HSDH Hidden-Markov Model search. (a) Number of subjects identified with putative 12β-HSDH genes present in their gut metagenomes. The metagenomes analyzed were from four distinct cohorts. (b) Distribution of microbial genomes with putative 12β-HSDH genes present across the four metagenomic studies
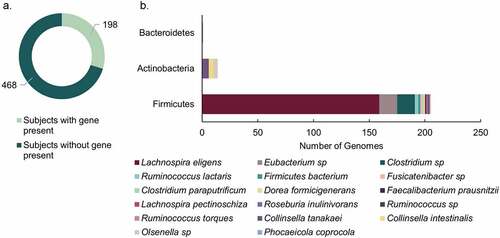
Figure 7. Maximum-likelihood phylogenetic analysis of regio- and stereospecific HSDHs. Clusters are shaded by function or marked as eukaryotic, as displayed in the legend. Sequences with experimentally determined activities are labeled with their function followed by organism and accession number. See Table S2 for sequence information