Figures & data
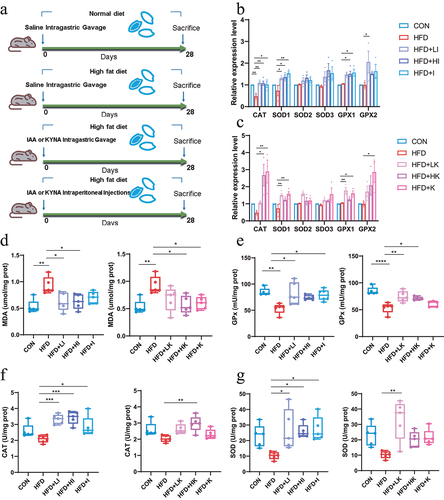
Figure 1. QA improved brain oxidative stress in mice subjected to HFD.
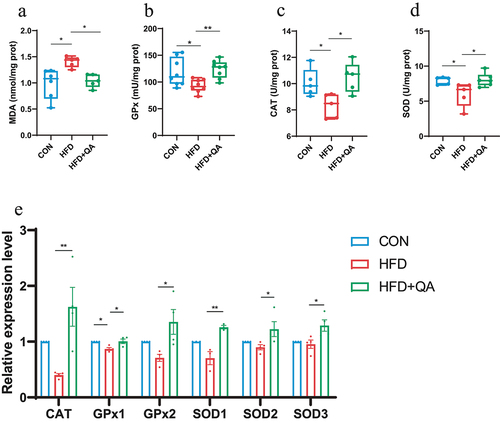
Figure 2. QA suppressed neuroinflammation and downregulated AD indicators in HFD mice.
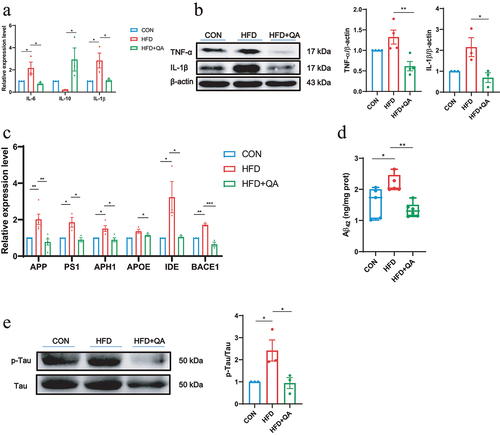
Figure 3. QA alleviated HFD-induced gut microbial dysbiosis.
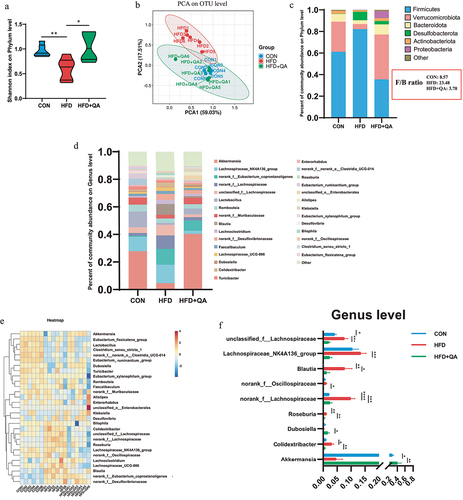
Figure 4. QA altered gut microbe-associated metabolites.
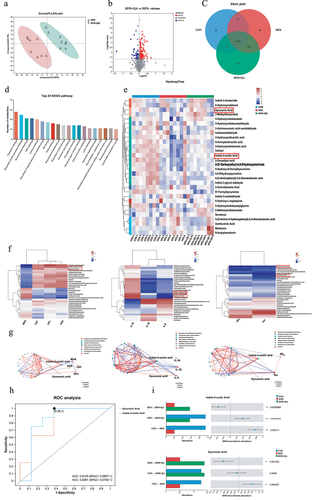
Figure 6. IAA and KYNA ameliorated inflammation of HFD mice.
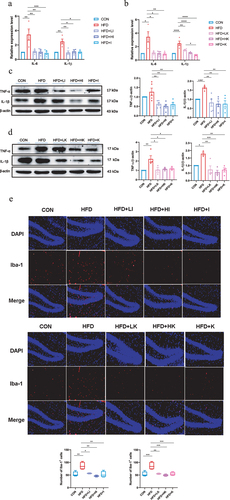
Figure 7. IAA and KYNA down-regulated AD-related indicators of HFD mice.
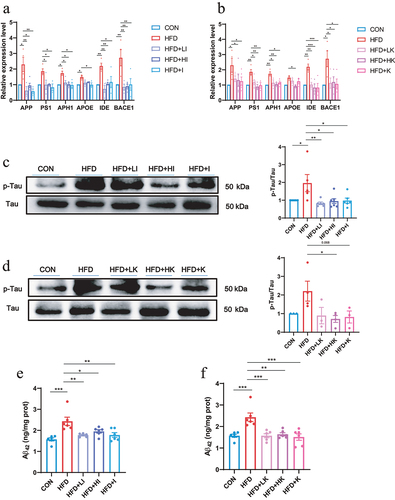
Figure 8. IAA altered the expression of genes in the brain of HFD mice.
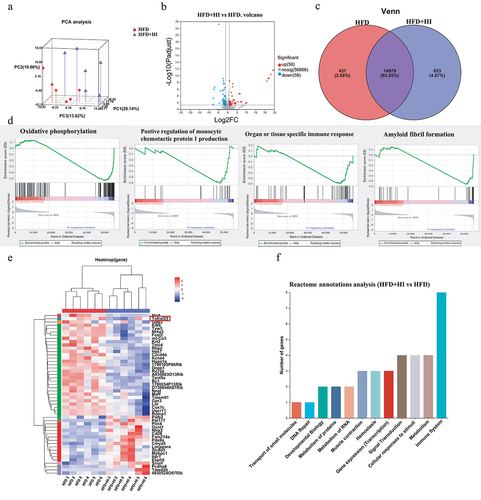
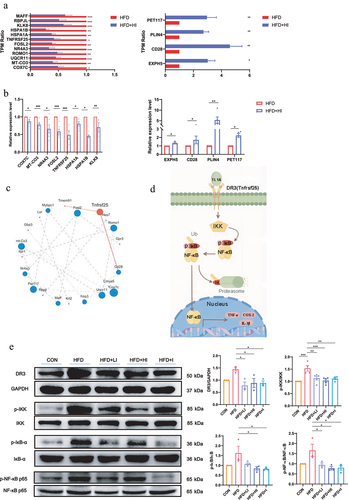
Figure 9. IAA-regulated neuroinflammation via the NF-κB signaling pathway.
Supplementary information.docx
Download MS Word (742 KB)WB.docx
Download MS Word (5.2 MB)Data availability statement
The data are available from the corresponding author on reasonable request.