Figures & data
Figure 1. Exon usage and sequence alignment of selected Tpm isoforms. (A, B) Exon usage of the (A) HMW and (B) LMW Tpm isoforms used in this study (adapted from Geeves et al., 2015). Shades of gray denote the different TPM genes. (C) Amino acid sequence comparison of Tpm isoforms. The amino acid sequences were aligned using ClustalX2. Exons are indicated above the sequence in the gray bars. The background color groups residues according to their physicochemical properties: orange = G; blue = hydrophobic (A, V, F, M, I, L) and C; blue-green = hydrophilic (Y, H); purple = acidic (D, E); red = basic (R, K); green = hydroxyl (S, T) and amide (N, Q).
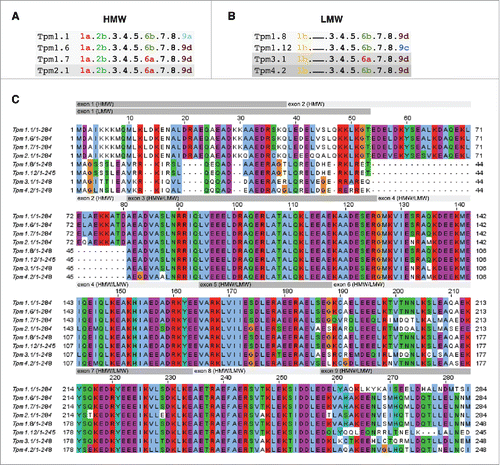
Figure 2. Expression and purification of Tpm4.2. (A) SDS PAGE with Coomassie Blue staining of total cell protein with and without induction of Tpm expression with IPTG. (B) Elution profile of the subtractive anion exchange chromatography step for removal of highly anionic contaminants such as nucleic acids. Tpm (monitored by UV light absorption at 280 nm and 230 nm) was collected in the flow through (peak between ∼30 and 100 ml elution volume). Bound nucleic acids (monitored by UV light absorption at 260 nm) was eluted with buffer B at ∼280 ml elution volume. (C, D) Elution profile (C) and SDS PAGE with Coomassie Blue staining (D) of the anion exchange chromatography step using a QFF column. Peak fractions containing highly enriched Tpm typically eluted at 35% buffer and were combined as indicated by the red bar. (E, F) Elution profile (E) and SDS PAGE with Coomassie Blue staining (F) of the chromatography using a ceramic hydroxyapatite column. Peak fractions containing pure Tpm were combined as indicated by the red bar. Note that in panels (B), (C) and (E), the green line represents concentration of buffer B (%) and the magenta line represents conductivity (mS/cm). (G) SDS PAGE with Coomassie Blue staining of all purified Tpm isoforms used in this study.
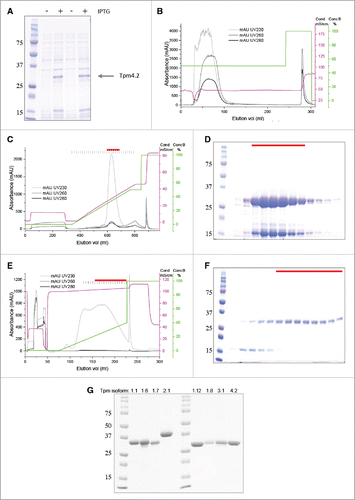
Figure 3. Binding curves of various Tpm isoforms to F-actin. (A) Binding of HMW Tpm isoforms to skeletal F-actin for Tpm1.1 (▪), Tpm1.6 (▴), Tpm 1.7 (•), and Tpm2.1 (▾); (B) Binding of LMW Tpm isoforms to skeletal F-actin for Tpm1.12 (□), Tpm 1.8 (Δ), Tpm3.1 (○), and Tpm4.2 (∇). (C) Comparison of the binding of selected HMW and LMW Tpm isoforms to either skeletal or cytoskeletal F-actin: Tpm1.1 to skeletal (▪), and cytoskeletal (▴) F-actin; Tpm3.1 to skeletal (○), and cytoskeletal (∇) F-actin. The best fit of the Hill equation to the data is shown as a solid line (skeletal F-actin) or dashed line (cytoskeletal F-actin). The apparent equilibrium dissociation constants (Kd(app)) and the Hill coefficients (h) are listed in . Buffer conditions: 150 mM NaCl, 10 mM Tris-HCl pH 7.5, 2 mM MgCl2, 0.5 mM DTT.
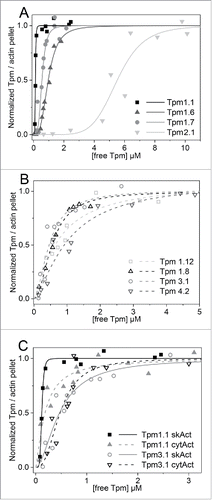
Table 1. Apparent equilibrium dissociation constants and Hill coefficients for binding of HMW and LMW tropomyosin isoforms to F-actin. Values for Kd(app) determined in previous studies for comparison: (a) Tpm1.1 with Ala-Ser extension: 0.18 μM,Citation25 0.20 μM,Citation23; (b) Tpm1.6: 0.82 μM,Citation35 0.32 μMCitation6; (c) Tpm2.1: >20 μMCitation25; (d) Tpm1.8: 0.27 μM,Citation35 0.09 μMCitation6; (e) Tpm1.12: 3.48 μM,Citation27 >10 μMCitation6; (f) Tpm3.1: 0.1 μMCitation27
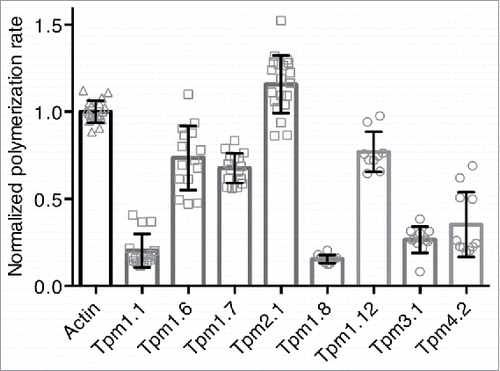
Figure 4. Effect of HMW and LMW Tpm isoforms on actin polymerization. Normalized polymerization rates of 2 μM G-actin (10 % pyrene labeled) in the presence of 0.5 μM unlabelled F-actin seeds and saturating concentrations of various Tpm isoforms. The Tpm concentration was 2.5 μM for all isoforms with exception of Tpm2.1 (7 μM). Data were obtained from at least 3 independent experiments and the errors are given as mean ± SD. Buffer conditions: 100 mM KCl, 2 mM MgCl2, 2 mM Tris-HCl pH 8, 0.1 mM CaCl2, 0.2 mM ATP, 0.5 mM DTT, 1 mM NaN3. The mean rate in the absence of Tpm was significantly different to the means in the presence of Tpm (one-way ANOVA with Tukey's test, p < 0.01 for actin vs. Tpm2.1, p < 0.001 for actin vs. Tpm1.12 and p < 0.0001 for actin vs. all other Tpm isoforms).