Figures & data
Figure 1. Cytoplasmic and nuclear mechanisms for lncRNAs. This summarizes just a few of the molecular mechanisms by which lncRNAs can function. This figure was created with BioRender.com.
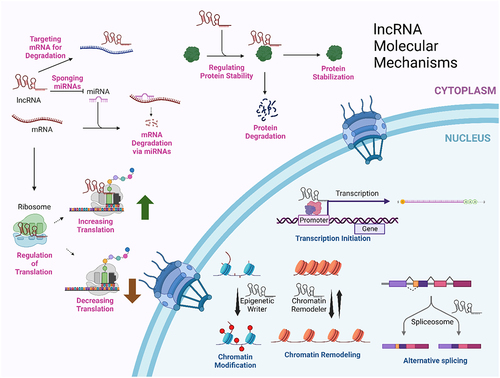
Table 1. Function of selected additional lncRNAs in cellular stress.
Figure 2. The RNP network of the nucleus is well preserved after chromatin removal. Shown are epon sections of CaSki cervical carcinoma cells before (a) and after (b) the isolation of a crosslink stabilized nuclear matrix and selectively stained for RNA by the EDTA- regressive procedure [Citation76] to visualize the RNP-network. The nuclear lamina (L) forms the periphery of the nucleus (panel A) and is retained in the nuclear matrix (panel B). The removal of chromatin after formaldehyde crosslinking does not substantially alter the structure or spatial distribution of the nuclear RNP network. (c) Higher magnification reveals well-preserved interchromatin granule clusters, enriched in RNA-splicing factors, in the RNP-network of the crosslink-stabilized nuclear matrix. The CaSki nuclear matrix in this panel was counterstained with an antibody recognizing the RNA-splicing factor SRm160 and a colloidal-gold-conjugated second antibody. The bar in panels a and B is 500 nm and in panel C the bar is 200 nm. This is from of Nickerson et al. [Citation82].
![Figure 2. The RNP network of the nucleus is well preserved after chromatin removal. Shown are epon sections of CaSki cervical carcinoma cells before (a) and after (b) the isolation of a crosslink stabilized nuclear matrix and selectively stained for RNA by the EDTA- regressive procedure [Citation76] to visualize the RNP-network. The nuclear lamina (L) forms the periphery of the nucleus (panel A) and is retained in the nuclear matrix (panel B). The removal of chromatin after formaldehyde crosslinking does not substantially alter the structure or spatial distribution of the nuclear RNP network. (c) Higher magnification reveals well-preserved interchromatin granule clusters, enriched in RNA-splicing factors, in the RNP-network of the crosslink-stabilized nuclear matrix. The CaSki nuclear matrix in this panel was counterstained with an antibody recognizing the RNA-splicing factor SRm160 and a colloidal-gold-conjugated second antibody. The bar in panels a and B is 500 nm and in panel C the bar is 200 nm. This is from Figure 4 of Nickerson et al. [Citation82].](/cms/asset/3769b107-ddf0-4afc-8d05-35e1ffad4139/kncl_a_2350180_f0002_b.gif)
Figure 3. The RNP network of a CaSki cell isolated after crosslink-stabilization and chromatin removal. When visualized by resinless section electron microscopy. (a) The RNP network or nuclear matrix consists of two parts, the nuclear lamina (L) and a network of structured fibers connected to the lamina and well distributed through the nuclear volume. The matrices of nucleoli (nu) remain and are connected to the fibers of the internal nuclear matrix. Three remnant nucleoli may be seen in this section. (b) Seen at higher magnification, the highly structured fibers of an internal RNP-netork or nuclear matrix are constructed on an underlying structure of 10-nm filaments, which occasionally branch. These are seen most clearly when, for short stretches, they are free of covering material (arrowheads). The bar shown in panel a represents 1 µM, and in panel B it is 100 nm. This is from of Nickerson et al., 1997 [Citation82].
![Figure 3. The RNP network of a CaSki cell isolated after crosslink-stabilization and chromatin removal. When visualized by resinless section electron microscopy. (a) The RNP network or nuclear matrix consists of two parts, the nuclear lamina (L) and a network of structured fibers connected to the lamina and well distributed through the nuclear volume. The matrices of nucleoli (nu) remain and are connected to the fibers of the internal nuclear matrix. Three remnant nucleoli may be seen in this section. (b) Seen at higher magnification, the highly structured fibers of an internal RNP-netork or nuclear matrix are constructed on an underlying structure of 10-nm filaments, which occasionally branch. These are seen most clearly when, for short stretches, they are free of covering material (arrowheads). The bar shown in panel a represents 1 µM, and in panel B it is 100 nm. This is from Figure 3 of Nickerson et al., 1997 [Citation82].](/cms/asset/ae249606-1623-499c-85f6-deb572fff7db/kncl_a_2350180_f0003_b.gif)
Figure 4. The RNA-containing nuclear matrix from a HeLa cell when isolated without a cross-linking step before the removal of chromatin. The structure is constructed on a branched network of 10-nm core filaments that resemble filaments that form in phase-separated condensates of RNP proteins [Citation169,Citation177–179]. This high magnification view shows connections between core filaments (arrow) and the nuclear lamina (L). The bar is 100nM [Citation180].
![Figure 4. The RNA-containing nuclear matrix from a HeLa cell when isolated without a cross-linking step before the removal of chromatin. The structure is constructed on a branched network of 10-nm core filaments that resemble filaments that form in phase-separated condensates of RNP proteins [Citation169,Citation177–179]. This high magnification view shows connections between core filaments (arrow) and the nuclear lamina (L). The bar is 100nM [Citation180].](/cms/asset/be897d40-5da6-45da-a0d1-09c45dcf51e7/kncl_a_2350180_f0004_b.gif)
Data availability statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
