Figures & data
Table 1. Primer pairs used to amplify the IGFBP-3 promoter region.
Figure 1. SNPs in the IGFBP-3 promoter region.
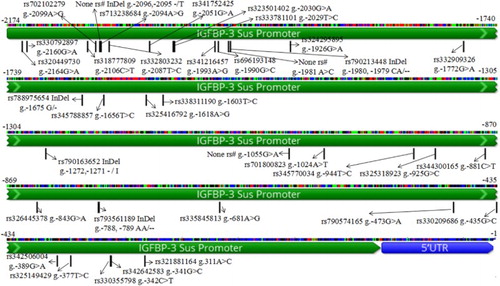
Table 2. Genetic diversity parameters of the IGFBP-3 promoter regiona in pig breeds.
Figure 2. Perfect LD structure and haplotypes for the 13 SNPs (r2 = 1) located in the IGFBP-3 promoter region.
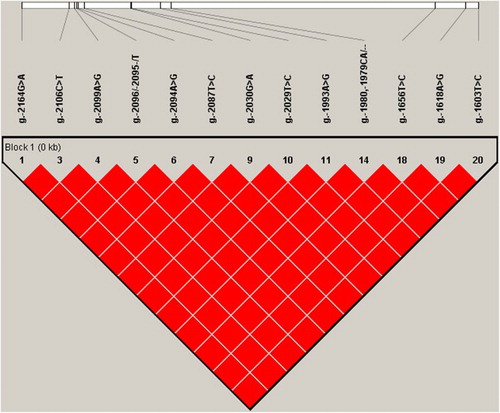
Table 3. Haplotypes of block 1 () and the frequency distributions in the pig breeds.
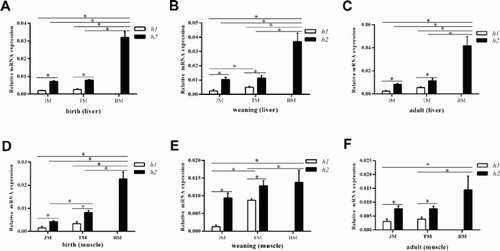
Figure 3. The haplotypes (h1, h2) of the promoter region are associated with the IGFBP-3 mRNA expression level in liver and muscle tissues of three pig breeds (JM, TM and BM).