Figures & data
Figure 1. Expression of TMEM230. (A) A scheme of TMEM230 WT and the tested mutants in isoforms 1 and 2. The locations of two putative transmembrane domains (TM), in R78L and PG5E (PGHPPHS) are indicated. (B) Western blot images of Flag-TMEM230 WT, -TMEM230 R78L or -TMEM230 PG5E expressed in SN4741, HEK293T and SH-SY5Y cells. (C) Fluorescence microscopic images of SN4741 cells expressing GFP, GFP-TMEM230 WT, -TMEM230 R78L or -TMEM230 PG5E.
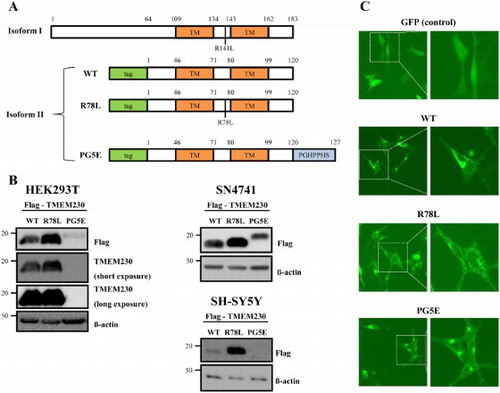
Figure 2. Stability of Flag-TMEM230 WT, -TMEM230 R78L or -TMEM230 PG5E proteins. (A) SN4741 cells were transfected with the indicated TMEM230 plasmids for 40 h and then treated with cycloheximide (CHX) for 0, 1, 3 and 8 h. Each TMEM230 protein level is analyzed by western blot. n = 3. (B) Relative level of TMEM230 proteins after cycloheximide treatment. (C) Western blot analysis of TMEM230 protein secreted in culture media. Alix and TSG101 were used as loading controls of media. A representative image is shown from three separate experiments (A, C).
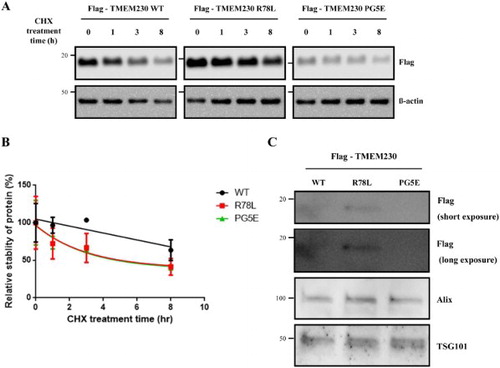
Figure 3. FACS analysis of SN4741 cells expressing GFP, GFP-TMEM230 WT or GFP-TMEM230 mutant proteins after treatment with or without MPP+. (A) Fluorescence images of cells expressing GFP proteins after MPP+ treatment. (B) Scatter plots showing the distribution of GFP and PI staining for each group. (C) Quantitative analysis of the percentage of dead cells by FACS analysis. The data were analyzed by two-way ANOVA followed by Tukey’s post-hoc test (n = 6). There was no significant difference among the cells transfected with the empty vector and TMEM genes and treated with DMSO. *p < 0.05, **p < 0.01, ****p < 0.0001.
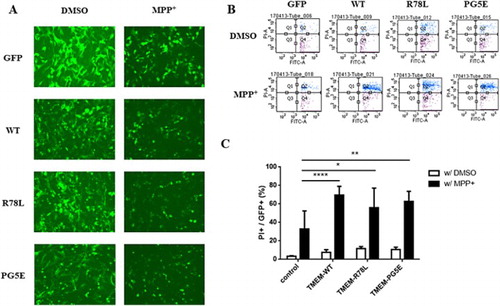
Figure 4. TMEM230 neither interacts with LRRK2 nor is a kinase substrate for LRRK2. A. LRRK2 kinase assay. Purified GST or GST-TMEM230 WT proteins were used as the substrate and incubated with [γ-32P]-ATP and recombinant GST-ΔN LRRK2 G2019S (GS) proteins in a LRRK2 kinase assay. CB: Coommasie blue; *: non-specific band. B. A GFP- trap assay for GFP-TMEM230 and LRRK2. HEK293T cells co-transfected with LRRK2 WT and GFP, GFP-TMEM230 WT, -TMEM230 R78L or -TMEM230 PG5E were used in the GFP-Trap assay. Immunoprecipitates were analyzed with anti-LRRK2 and anti-GFP antibodies.
![Figure 4. TMEM230 neither interacts with LRRK2 nor is a kinase substrate for LRRK2. A. LRRK2 kinase assay. Purified GST or GST-TMEM230 WT proteins were used as the substrate and incubated with [γ-32P]-ATP and recombinant GST-ΔN LRRK2 G2019S (GS) proteins in a LRRK2 kinase assay. CB: Coommasie blue; *: non-specific band. B. A GFP- trap assay for GFP-TMEM230 and LRRK2. HEK293T cells co-transfected with LRRK2 WT and GFP, GFP-TMEM230 WT, -TMEM230 R78L or -TMEM230 PG5E were used in the GFP-Trap assay. Immunoprecipitates were analyzed with anti-LRRK2 and anti-GFP antibodies.](/cms/asset/92c4cba0-3051-4f9f-8e0e-215fe7313edf/tacs_a_1453545_f0004_b.gif)
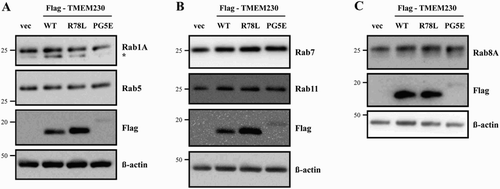
Figure 5. Effect of TMEM230 expression on Rab protein levels. Total lysates of SN4741 cells expressing Flag-TMEM230 WT, -TMEM230 R78L or -TMEM230 PG5E were prepared, loaded into three separate gels (A, B and C) and analyzed by the indicated antibodies by western blotting (n = 3). vec: control vector. *: non-specific band.