Figures & data
Table 1. Oligonucleotides primers for real-time PCR assays used for bacteria quantification in biofilms and for transcription analysis of P. gingivalis virulence-associated genes.
Figure 1. Effect of probiotics cell-free supernatants (CFS) diluted at 1:2.5 on P. gingivalis W83 (a1, mono-species; a2, multi-species) and ATCC 33277 (b1, mono-species; b2, multi-species) biofilm biomass, represented by the OD490nm of the standard biofilm dye. Groups: Neg. control- Negative control represents non-inoculated medium; Control- CFS-free positive controls of P. gingivalis mono- and multi-species biofilms (So – S. oralis and Sg – S. gordonii), and experimental group with CFS of: LA5 – L. acidophilus LA5, HN001 – L. rhamnosus HN001, DSM – L. reuteri DSM 17938, 1101A – B. breve 1101A, 1191A – B. pseudolongum 1191A and 1622A – B. bifidum 1622A. Experiments were conducted in triplicate. (*) Statistically significant difference when compared to respective positive controls using One-way ANOVA with post hoc Tukey’s multiple comparisons (p < 0.05).
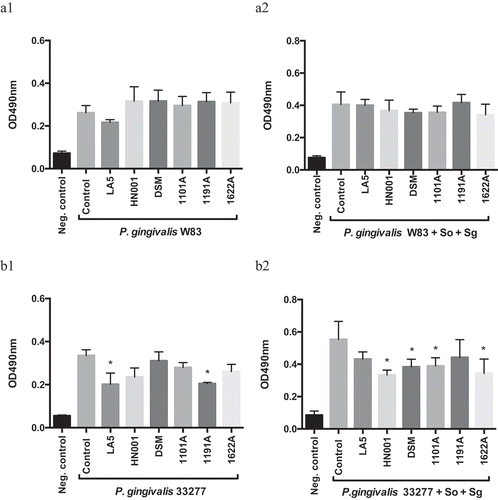
Figure 2. Effect of probiotics cell-free supernatants (CFS of LA5 – L. acidophilus LA5, HN001 – L. rhamnosus HN001, DSM – L. reuteri DSM 17938, 1101A – B. breve 1101A, 1191A – B. pseudolongum 1191A and 1622A – B. bifidum 1622A) on the relative abundance of P. gingivalis W83 (a) or ATCC 33277 (b) and the initial colonizers S. oralis and S. gordonii in multi-species biofilms, represented as the mean percentage of each bacteria determined by qPCR. (*) Significant difference in P. gingivalis counts when compared to respective positive controls using One-way ANOVA with post hoc Tukey’s multiple comparisons (p < 0.05).
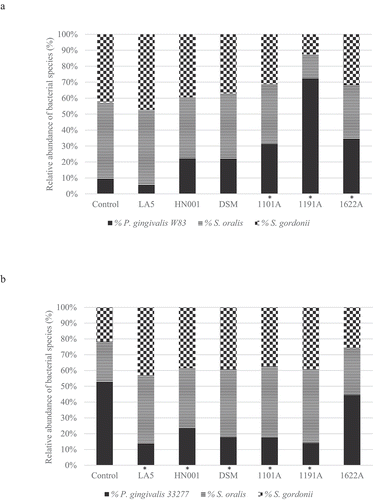
Figure 3. Effect of living Lactobacillus sp (L. acidophilus LA5, L. rhamnosus HN001, L. reuteri DSM 17938) and Bifidobacterium sp (B. breve 1101A, B. pseudolongum 1191A and B. bifidum 1622A) on the relative abundance of P. gingivalis W83 (a) or ATCC 33277 (b) and the initial colonizers S. oralis and S. gordonii after cell-to-cell interaction, represented as the mean percentage of each bacteria determined by qPCR (*). Significant difference in P. gingivalis abundance, when compared to respective positive controls using One-way ANOVA with post hoc Tukey’s multiple comparisons (p <0.05).
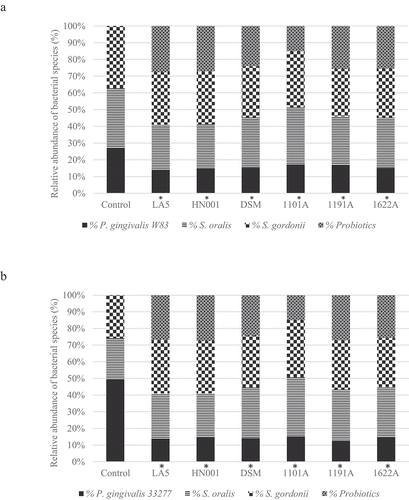
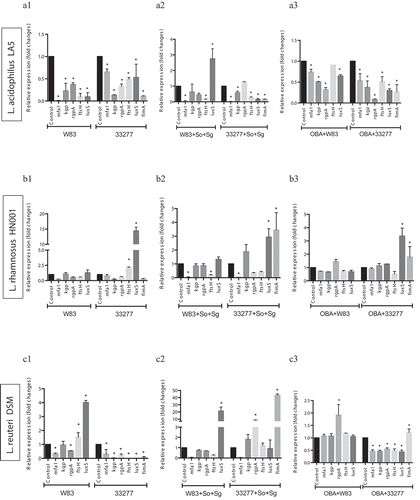
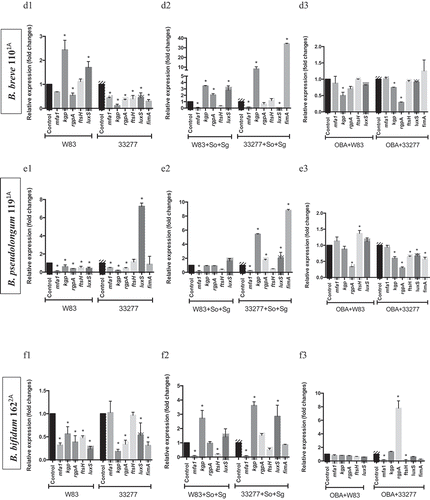
Figure 4. Effect of probiotics on the relative transcription of P. gingivalis encoding virulence genes (mfa1 – minor fimbriae; fimA – major fimbriae; kgp – lysine gingipain; rgpA – arginine gingipain; ftsH – metalloproteinase, and luxS – quorum sensing components), determined by RT-qPCR. P. gingivalis strains W83 and ATCC 33277 biofilms formed in BHIHM broth added with the supernatant of cultures of probiotics (a1 and a2 - L. acidophilus LA5, b1 and b2 – L. rhamnosus HN001, c1 and c2 – L. reuteri DSM 17938, d1 and d2 – B. breve 1101A, e1 and e2 – B. pseudolongum 1191A and f1 and f2 – B. bifidum 1622A) diluted to 1:2.5, in mono-species (a1, b1, c1, d1, 1 and f1) and multi-species (a2, b2, c2, d2, e2 and f2), and infecting OBA-9 GECs with probiotic co-infection at a MOI of 1:1,000 (a3 - L. acidophilus LA5, b3 – L. rhamnosus HN001, c3 – L. reuteri DSM 17938, d3 – B. breve 1101A, e3 – B. pseudolongum 1191A and f3 – B. bifidum 1622A). Data are expressed as fold changes in relation to positive control conditions (biofilms without probiotic cell-free supernatants or GECs without probiotic bacteria), after normalization to the endogenous control gene 16SrRNA. (*) Significant difference when compared to respective positive controls using One-way ANOVA with post hoc Tukey’s multiple comparisons (p < 0.05).