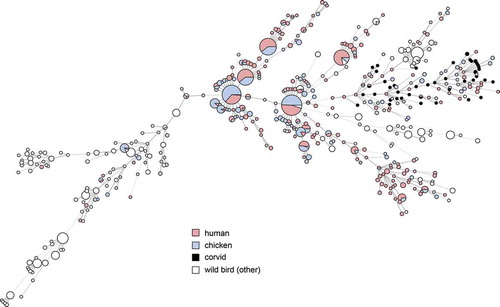
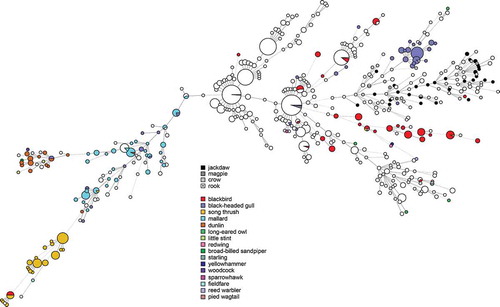
Figures & data
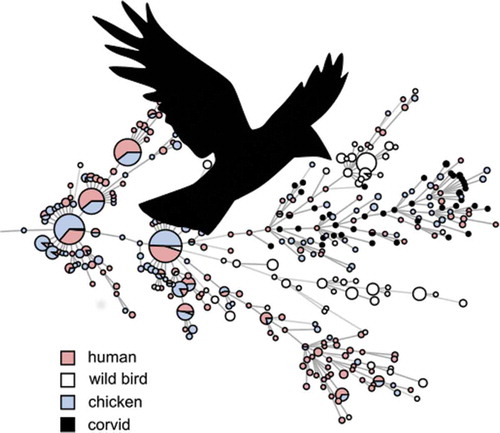
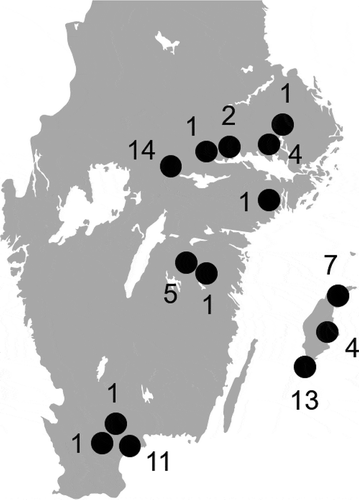
Figure 1. Approximate sampling locations (black dots) and the number of birds sampled at each location.
Data availability statement
Data from high-throughput sequencing of the recovered bacterial isolates have been deposited in ENA (https://ebi.ac.uk/ena) under the accession number PRJEB34530. MLST profiles have been deposited in the pubMLST database (https://pubmlst.org) with isolate identities SVAcorv1-SVAcorv32, SVAcorv34-SVAcorv40.