Figures & data
Table 1. Laboratory and clinically derived daptomycin-exposed Staphylococcus aureus strains used in this study
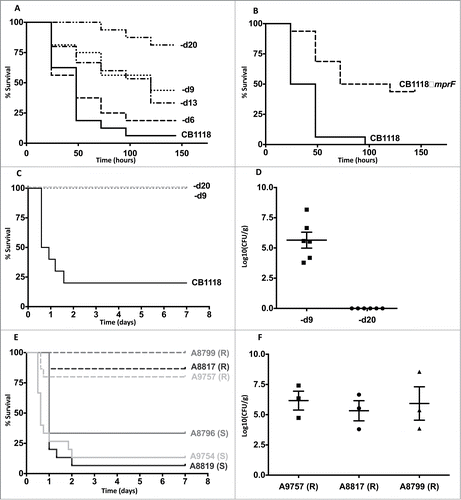
Figure 1. Daptomycin resistance in S. aureus correlates with altered virulence and in vivo persistence. (A) G. mellonella infection of a laboratory-derived series of isolates with incremental increases in daptomycin MIC was performed (n = 16 for each strain). For clarity, CB1618-d6, CB1618-d9, CB1618-d13, CB1618-d20 are represented by d6, d9, d13 and d20 respectively. Virulence attenuation was observed for CB1618-d9 when compared to CB1618-d6 (P < 0.01) and CB1618-d20 when compared with CB1618-d13 (P < 0.01). No significant virulence attenuation was observed for CB1618-d6 (P = 0.44) and CB1618-d13 (P = 0.70) when compared to their respective progenitor strains. (B) An mprF deletion strain (CB1118ΔmprF) produced significantly less killing of G. mellonella when compared to its progenitor (P < 0.001, n = 16 for each strain). (C) CB1618-d9 and CB1618-d20 were attenuated for virulence in a murine septicaemia model (P < 0.001, n = 10 for each strain). (D) CB1618-d9 was capable of in vivo persistence as determined by bacterial densities in the kidneys of mice 7-days post infection. In contrast, bacterial burden was not observed in the kidneys of mice infected with CB1618-d20. (E) Virulence of 3 DAP-exposed clinical pairs was assessed using a murine septicaemia model. The daptomycin-resistant (R) isolates were significantly attenuated for virulence compared to their susceptible progenitors (S) (P < 0.001, n = 15 for each strain) and (F) were persistent in the kidneys of infected mice out to 7-days post-infection.