Figures & data
Figure 1. Circular representation of pEntYN10. The innermost circle represents G + C content (% of 1,450 bp window length). Greater and less than 50% are colored respectively as purple and green. The outer circles represent annotated CDSs with their strand orientation (outer – forward; inner – reverse). Colors show their putative categories of function as follows: virulence and cell adhesion, red; IS, pink; replication and plasmid stability, green; other functions, blue. Virulence and cell adhesion related CDSs were assigned by their names.
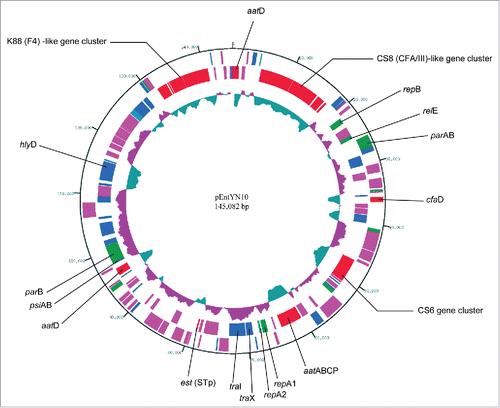
Table 1. Characteristics of RepFIIA plasmids from human ETEC strains
Figure 2. Stability of pEntYN10 during passages was verified during 7 d by ΔΔCt comparison between genes on plasmid (est)/chromosome (gyrB) using realtime PCR. The Y-axis represents the difference of Ct value (ΔCt) between est and gyrB of respective strains. Asterisks denote statistically significant difference (*P < 0.001) by Student's t-test between ETEC strain O169:H41 (YN10) and positive control (H10407).
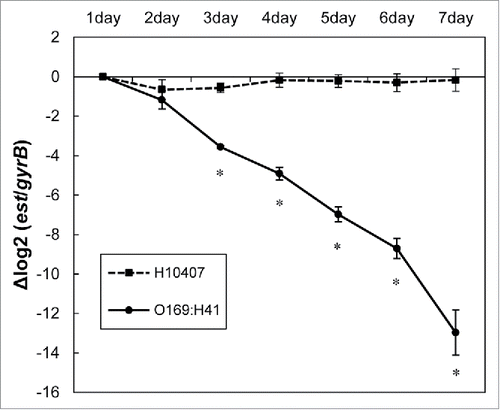
Figure 3. Linear comparison of adhesion related gene clusters between pEntYN10 and other plasmids of ETEC strains reported previously. The gene clusters were retrieved from the GenBank database: (A) CS6 [accession ID:U04846]; (B) CFA/III [accession ID:AB049751]; (C) K88 gene clusters for comparison were retrieved from the complete sequence of plasmid pUMNK88 [accession ID:CP002730]. The length scales are shown in respective bars. Genes coding fimbrial structure proteins are shown as greyed. They were analyzed in detail for this study.
![Figure 3. Linear comparison of adhesion related gene clusters between pEntYN10 and other plasmids of ETEC strains reported previously. The gene clusters were retrieved from the GenBank database: (A) CS6 [accession ID:U04846]; (B) CFA/III [accession ID:AB049751]; (C) K88 gene clusters for comparison were retrieved from the complete sequence of plasmid pUMNK88 [accession ID:CP002730]. The length scales are shown in respective bars. Genes coding fimbrial structure proteins are shown as greyed. They were analyzed in detail for this study.](/cms/asset/1c7eb716-4029-49ee-a5f1-0c54c8782bc5/kvir_a_1094606_f0003_b.gif)
Figure 4. Light micrograph of HEp-2 cells infected by ETEC strains (A). Scale bars, 10 μm. The number of adhered bacteria was counted quantitatively for each strain (B). Asterisks denote statistically significant difference (*P < 0.001) by Student's t-test between ETEC strain O169:H41 (YN10) and other strains.
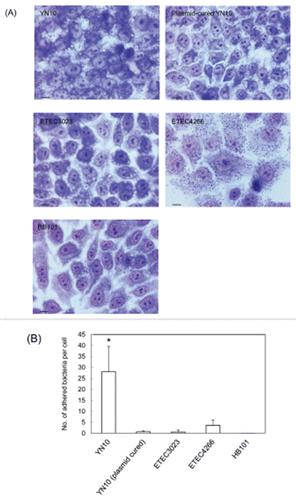
Table 2. Predicted CDSs on pEntYN10 related to cell adhesion
Figure 5. Sequence variation of CssA and CssB subunit of CS6. (A) CssA. Blue labels are referential amino acids for the type AI, yellow for type AII, and green for type AIII. Red labels differ from the reference amino acids. (B) CssB. Blue labels are referential amino acids for type BI, and yellow labels are for the type BII. Numbers refer to amino acid residues of the reference sequences.
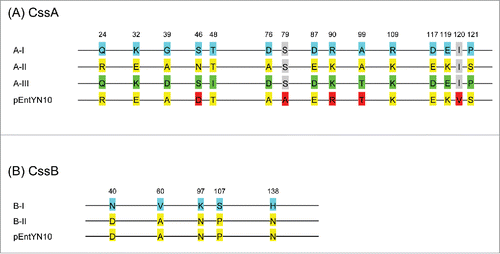
Figure 6. Comparison of CofA between pEntYN10 and ETEC 260-1 [BAB62897] and TcpA from V. cholerae O1 [AAK20785]. (A) A phylogenetic tree based on Neighbor Joining method. (B) Amino acid sequence alignment of the 3 proteins by CLUSTAL 2.1. Highly conserved residues responsible for the stable structure of pili of V. cholerae (Met26, Glu30, Arg51, Leu98, and Glu108) are shaded.
![Figure 6. Comparison of CofA between pEntYN10 and ETEC 260-1 [BAB62897] and TcpA from V. cholerae O1 [AAK20785]. (A) A phylogenetic tree based on Neighbor Joining method. (B) Amino acid sequence alignment of the 3 proteins by CLUSTAL 2.1. Highly conserved residues responsible for the stable structure of pili of V. cholerae (Met26, Glu30, Arg51, Leu98, and Glu108) are shaded.](/cms/asset/e2e803b0-f702-45c8-9623-76b6467cc704/kvir_a_1094606_f0006_b.gif)
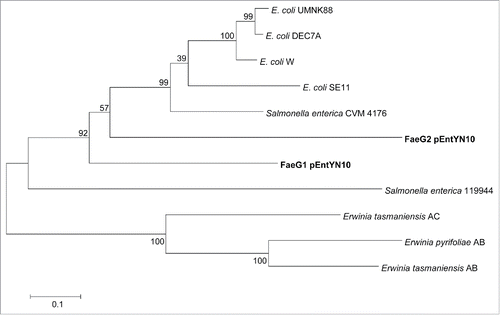
Figure 7. Phylogenetic tree based on amino acid sequences of FaeG. Numbers on nodes of the phylogenetic tree correspond to bootstrap values.