Figures & data
Figure 1. (A) LDH levels was measured in THP-1 monolayers that were infected with the Mtb wild-type and transposon mutants at a MOI of 10 bacterium to 1 cell, and the percentage of necrosis was calculated according to the manufacturer's protocol. The MOI used was 10. **, p < 0.01, the significance of differences between Mtb transposon mutants and the wild-type. (B) Survival assay for Mtb necrosis deficient mutants in vitro. THP-1 cells were infected with Mtb wild-type and 7 NDMs for 1h and 5 days; CFUs were recorded from lysed macrophages as well as from the supernatants of culture medium at day 5 post-infection. Results represent means ± standard error of the mean of 2 independent experiments. **, p < 0.01, the significance of differences between Mtb transposon mutants and the wild-type.
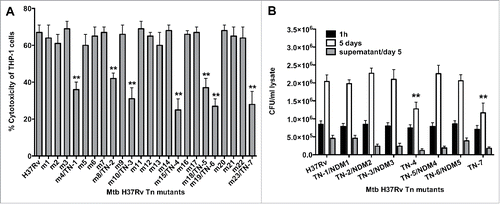
Figure 2. (A) Fluorescence micrographs of THP-1 cells at 7 days post-infection with the Mtb wild-type and NDMs. Macrophage monolayers were stained with both Annexin V-FITC and propidium iodide to visualize the early and late stage of apoptosis and/or necrosis of THP-1 cells, respectively. The early apoptotic THP-1 cells with intact membranes are seen in green; Late apoptotic cells that have already lost their membrane integrity are seen in orange (or orange to red) with green membrane fragments; Totally destroyed necrotic cells are seen in red. (B) The percentage of necrosis quantified in 2 hundred cells that was infected with either the Mtb wild-type or NDM transposon mutants. Results represent means ± standard error of the mean of 3 independent experiments. **, p < 0.01 and *, p < 0.05, the significance of differences between Mtb NDMs and the wild-type.
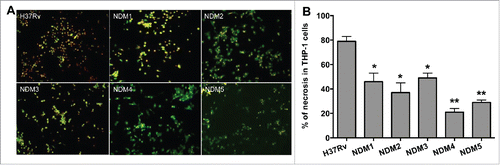
Figure 3. (A) Immunofluorescence analysis of mitochondrial transmembrane disruption after Mtb wild-type and NDMs infection compared to uninfected cells and staurosporine treatment (positive control). Red scattered staining in THP-1 cells indicate undamaged mitochondria; however, cells with altered mitochondrial membrane stains green as the dye accumulates in the cytoplasm and remains in its monomeric form. (B) The percentage of necrosis quantified in 2 hundred cells that was infected with either the Mtb wild-type or NDMs at MOI of 10. **, p < 0.01 and *, p < 0.05, the significance of differences between NDMs and the wild-type.
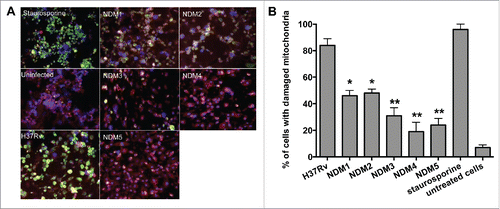
Figure 4. (A) Schematic organization of the M. tuberculosis RD1 region containing the PPE68 gene. (B) His-tagged pull-down assay of Mtb proteins interacting with PPE68. The overexpressed PPE68 protein in induced (lane 1) and uninduced (lane 2) E.coli cells is visualized with anti-His antibody. Lane 3, protein marker; lane 4, the pull-down control using the uninduced E.coli cell lysate; lane 5, Mtb bound proteins to PPE68. The membrane was visualized with Li-Cor Odyssey imaging system. (C) The interactions between PPE68 and Rv0462/lpdC (1), Rv0652/rplL (2), Rv1093/glyA1 (3), Rv1388/mihF (4), Rv1837c/glcB (5), Rv2220/glnA1 (6) and Rv2626c (7) were confirmed using a yeast 2-hybrid system. To test the specificity of PPE68 interaction to target Mtb proteins, the yeast reporter strain containing the bait pGBKT7:PPE4 and pGBKT7:PPE27 constructs were screened against the selected Mtb gene constructs in pGADT7 vector; The yeast zygotes that grew on quadruple dropout medium SD/–Ade/–His/–Leu/–Trp supplemented with X-a-Gal and Aureobasidin A (QDO/X/A) and turned blue in color were considered positive. The growth of the yeast cells on the –Trp/Lew medium was used as a control that diploid yeast cells contained both bait and pray constructs.
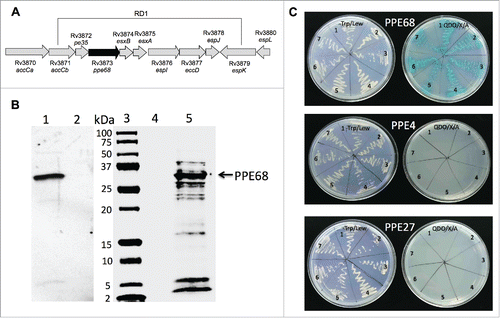
Table 1. PPE68 interacting Mtb proteins.
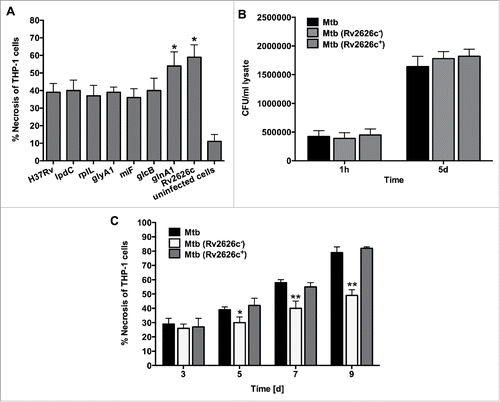
Figure 5. (A) Necrosis in THP-1 cells that was infected with the Mtb wild-type and overexpressed clones of lpdC, rplL, glyA1, mihF, glcB, glnA1 and Rv2626c was measured by the LDH activity assay. The MOI used was 10. Results represent means ± standard error of the mean of 3 independent experiments. *, p < 0.05, the significance of differences between Mtb overexpressed clones and the wild-type. (B) Infection and survival assay for Mtb knockout and complemented clone of Rv2626c in vitro. THP-1 cells were infected with Mtb wild-type, Mtb (Rv2626c−) and Mtb (Rv2626c+) clones and CFUs were recorded at 1 h and 5 days post-infection. Results that represent means ± standard error of the mean of 2 independent experiments indicate that Mtb knockout and complemented clones invade and survive in macrophages similarly to the wild-type strain. (C) The Mtb (Rv2626c−) infection of THP-1 cells demonstrates significantly less necrosis when compared to Mtb wild-type and Mtb (Rv2626c+) infection. Cells were infected with an MOI of 10. **, p < 0.01 and *, p < 0.05, the significance of differences between Mtb (Rv2626c−) and the wild-type and complemented clone.