Figures & data
Figure 1. Inhibition of Pb14-3-3 expression using aRNA and A. tumefaciens-mediated transformation. (A) tDNA (T-DNA) inserted into P. brasiliensis by ATMT to silence Pb14-3-3. The anti-sense oligonucleotides were produced based on the Pb18 (PbWT) genomic sequence, as detailed in the Materials and Methods section. The sequence was cloned under the control of CBP-1 promoter (calcium-binding protein). The construct was sub-cloned into T-DNA region of the binary vector pUR5750 harboring hygromycin B phosphotransferase (HPH) driven by the glyceraldehyde-3-phosphate of Aspergillus nidulans (PGPDA) and with terminator (TTRPC) as described previously.Citation31 (B) PCR fragments amplified with an HPH-specific primer yielded a 1,000 bp internal fragment. Pb14-3-3 aRNA, PbWT and PbEV were used as the templates. MW, DNA molecular marker. C) Pb14-3-3 expression levels in PbWT, PbEV and Pb14-3-3 aRNA after 1 month and over 12 months of subculture. Gene expression levels obtained by RT-qPCR were normalized to the internal control TUB2; ***p < 0.0001 when compared with PbWT and PbEV; D) SDS-PAGE gel of total proteins of PbWT and Pb14-3-3 aRNA; E) Immunoblots of the total proteins tested with the anti-14-3-3 monoclonal antibody and anti-enolase (used as an internal control) and quantified using ImageJ software; p = 0.0021.
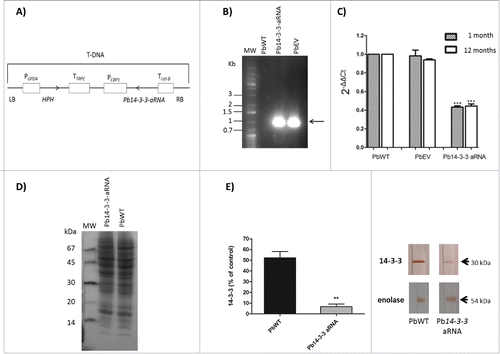
Table 1. Percentage of budding cells and number of buds by cell exhibited by the isolates.
Figure 2. Knocking-down Pb14-3-3 in P. brasiliensis alters yeast cell size. Analysis of major diameter size of mother cells of PbWT, PbEV and Pb14-3-3 aRNA isolates. (***p < 0.0001); n = 100 mother cells.

Figure 3. Silencing of Pb14-3-3 leads to distinct P. brasiliensis yeast cell morphology. Microscopic evaluation of PbWT and PbEV yeast cells and yeast cells generated from the Pb14-3-3 aRNA strain using CalcoFluor; magnification 40X. White bars correspond to 20 μm. White arrows indicate yeast cells showing elongation and the presence of filamentation.
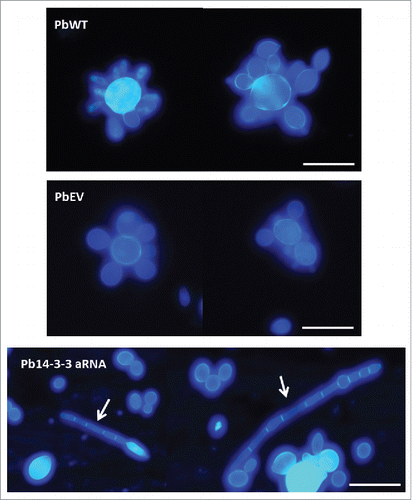
Figure 4. Influence of Pb14-3-3 in the temperature morphological transition. Mycelia or yeast were induced to transform by changing the incubation temperature from 25°C to 37°C or from 37°C to 25°C. Morphological units were classified as described by Nunes et al., 2005Citation83: (A) Yeast (multiple budding), (B) Transforming yeast (yeast with neck elongation), (C) Differentiating hyphae (chlamydospore-like cells) and (D) hyphae. (E) Yeast-Mycelium transition at 25°C. (F) Mycelium-Yeast transition at 37°C. A total of 300 cells were counted at each time, and the results are shown as the mean.
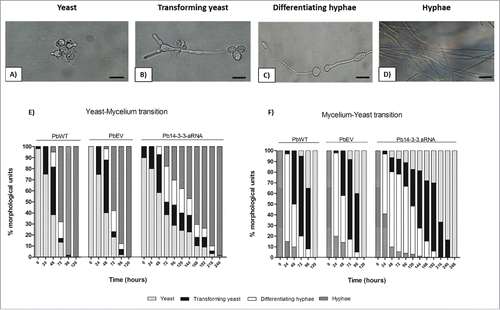
Figure 5. Down-regulation of Pb14-3-3 does not affect cell viability or vitality of P. brasiliensis yeast cell. P. brasiliensis isolates PbWT, PbEV and Pb14-3-3 aRNA were evaluated by (A) cell counts in a Neubauer chamber, (B) a colorimetric XTT reduction assay and (C) Trypan blue exclusion test at different time points.
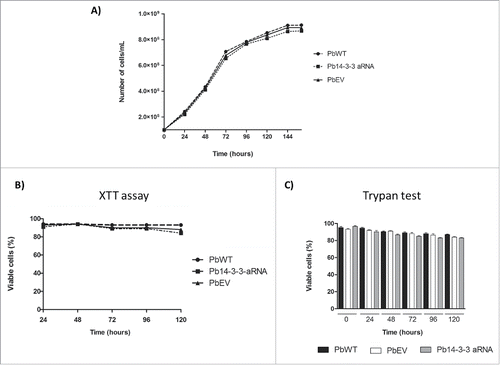
Figure 6. Interaction of Pb14-3-3 aRNA yeast cells with pneumocytes is affected during the early stage of infection. The interaction was assessed by indirect immunofluorescence and analyzed by flow cytometry at different time points. *p < 0.05 when comparing PbWT and PbEV with Pb14-3-3 aRNA at 2 and 5 hours after infection.
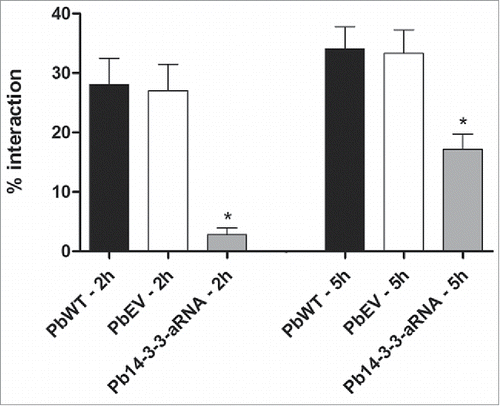
Figure 7. Binding of the Pb14-3-3 mutant to ECM is reduced compared to the PbWT. Binding yeast cells of the strains PbWT, PbEV and Pb14-3-3 aRNA to the ECM components laminin, fibronectin, and type I and type IV collagen (10 µg mL−1). The interaction was determined by ELISA with peroxidase. *p < 0.05 when Pb14-3-3 aRNA was compared to PbWT and PbEV for laminin and fibronectin.
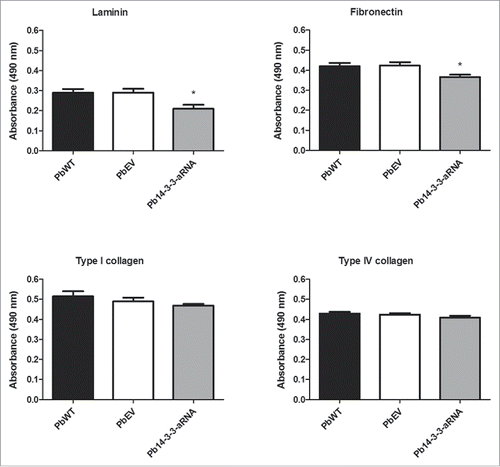
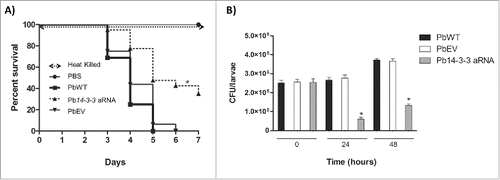
Figure 8. Silencing of Pb14-3-3 decreases the virulence of P. brasiliensis in a Galleria mellonella infection model. (A) Representative survival curves of an experimental infection performed in G. mellonella with PBS; 5×106 PbWT, PbEV, Pb14-3-3 aRNA and heat-killed yeast cells; p < 0.0001 when Pb14-3-3 aRNA were compared with PbWT and PbEV and B) Number of CFUs in larvae sacrificed and homogenized at 0, 24 and 48 h after infection (*p < 0.05 when compared Pb14-3-3 aRNA with PbWT and PbEV).