Figures & data
Figure 1. CaeA prevents activation of executioner caspase 7. HEK293 cells stably expressing GFP or GFP-CaeA were exposed to UV-light (200 J/m2 or 800 J/m2) and incubated for 6 h at 37°C in 5% CO2. Proteins were separated by SDS-PAGE, transferred to a PVDF membrane and probed with antibodies against (a) Bcl-2, Bcl-xL, Mcl-1 and actin as loading control or (b) cleaved caspase 9, cleaved caspase 7, cleaved PARP and actin as loading control. The result of one representative experiment out of 3 independent experiments with similar results is shown. (c) HeLa cells were transiently transfected with plasmids encoding GFP as control or GFP-CaeA followed by treatment with TNF (20 ng/ml) and cycloheximide (6 µg/ml) for 5 h at 37°C in 5% CO2. The cells were fixed, permeabilized and the nuclei were stained with DAPI. The nuclear morphology of GFP-expressing cells was scored. Data represent average values ± SD of 100 nuclei counted per sample from GFP-expressing cells from 3 independent experiments. (d) HeLa-Fas cells were transiently transfected with plasmids encoding GFP as control or GFP-CaeA followed by treatment with 0.125 µg/ml anti-Fas IgG for 5 h. The cells were fixed, permeabilized and the nuclei were stained with DAPI. The nuclear morphology of GFP-expressing cells was scored. Data represent average values ± SD of 100 nuclei counted per sample from GFP-expressing cells from 3 independent experiments.
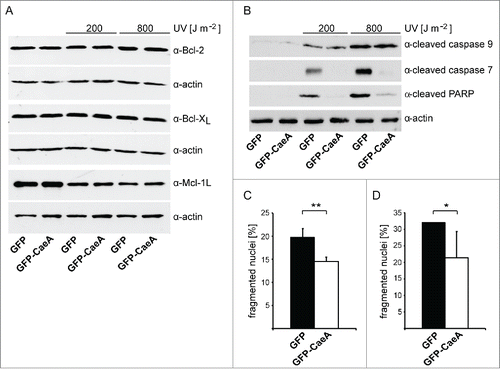
Figure 2. CaeA leads to survivin up-regulation. HEK293 cells stably expressing GFP or GFP-CaeA were exposed to UV-light (200 J/m2 or 800 J/m2) and incubated for 6 h. (a) Proteins were separated by SDS-PAGE, transferred to a PVDF membrane and probed with antibodies against survivin, XIAP and actin as loading control. The result of one representative experiment out of 3 independent experiments with similar results is shown. (b) Densitometric analysis of the survivin/actin ratio was performed using ImageJ. The data shown represent average values ± SD from at least 3 independent experiments. *P < 0.05 ***P < 0.001
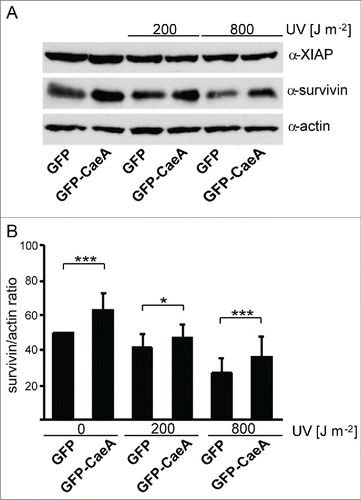
Figure 3. Survivin knock-down does not affect CaeA-induced apoptosis-inhibition. HEK293 cells stably expressing GFP or GFP-CaeA were transfected with 50nM non-targeting siRNA (−) or human survivin siRNA (+). (a) 48 hours post-transfection total isolated RNAs were reverse transcribed using SuperScript II reverse transcriptase according to the manufacturer's protocol and a qRT-PCR was performed with oligonucleotides specific for survivin and hpbgd as a house keeping gene. The ΔΔCt values were calculated for the fold difference of survivin siRNA-treated cells to non-targeting siRNA-treated cells using the 2−ΔΔ Ct method. (b) 48 hours post-transfection cells were treated with or without UV light (800 J/m2) and incubated for 6h at 37°C in 5% CO2. Proteins were separated by SDS-PAGE, transferred to a PVDF membrane and probed with antibodies against cleaved PARP, survivin and actin as loading control. The result of one representative experiment out of 3 independent experiments with similar results is shown.
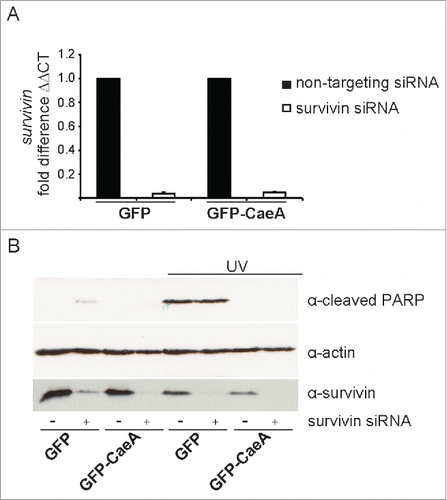
Figure 4. Genomic plasticity in the caeA gene. Genomic DNA of 25 C. burnetii isolates was prepared, the gene caeA and flanking regions was amplified and sequenced, and a multiple sequence alignment was performed. The illustrated alignment includes 63 bp upstream of the start codon (underlined) and the first 182–240 bp of the caeA open reading frame. Nucleotides matching in all isolates are marked by a dot and single base deletions are indicated by a dash in a red box. Non-displayed regions (positions 7–48 and 62–129) are identical in all isolates. Within the sequence of caeA each isolate contains a GAA AAG/A (EK) motif with a different number (3 to 13) of repetitions (blue box). Strains were grouped into 7 clusters (I to VII) according to their EK-repetition motif profile. The first base deletion (nt∼55) results in a frame shift and premature stop codon ‘TAA’ at bp 61 to 63, the second base deletion (nt∼136) similarly provokes a ‘TAA’ stop codon immediately downstream of the GAA AAG/A motif.
Figure 5. The EK repetition motif is required for apoptosis inhibition. (a) HEK293 cells and HEK293 cells stably expressing GFP, GFP-CaeA and GFP-CaeAΔEK were analyzed by flow cytometry. One representative experiment out of at least 3 independent experiments with similar results is depicted. (b and c) HEK293 cells stably expressing GFP, GFP-CaeA or GFP-CaeAΔEK were exposed to the UV-light intensities indicated and incubated for 6 h at 37°C in 5% CO2. Proteins were separated by SDS-PAGE, transferred to a PVDF membrane and probed with antibodies against (b) cleaved caspase 7, cleaved PARP (c) and actin as loading control. The result of one representative experiment out of at least 3 independent experiments with similar results is shown.
Figure 6. Deletion of the EK repetition motif does not alter intracellular localization of CaeA. Representative immunofluorescence micrographs show CHO-FcR cells transiently transfected with GFP, GFP-CaeA and GFP-CaeAΔEK. The cells were treated with or without 1 µM staurosporine for 6 h at 37°C in 5% CO2 followed by fixation, permeabilization and staining of the nuclei with DAPI (blue).
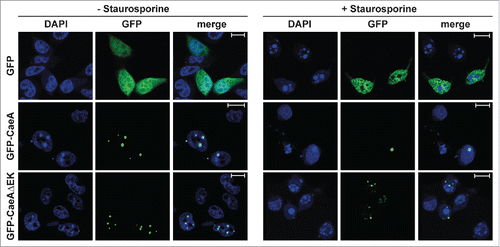
Figure 7. Four EK repeats are sufficient for inhibition of apoptosis. CHO-FcR cells transiently transfected with GFP, GFP-CaeA, GFP-CaeAΔEK, GFP-CaeAΔEK+2EK and GFP-CaeAΔEK+4EK were treated with 1µM staurosporine for 6 h at 37°C in 5% CO2. The cells were fixed, permeabilized and the nuclei were stained with DAPI. The nuclear morphology of GFP-expressing cells was scored. Data represent average values ± SD of 100 nuclei counted per sample from GFP-expressing cells from 5 independent experiments. **P < 0.01, ***P < 0.001.
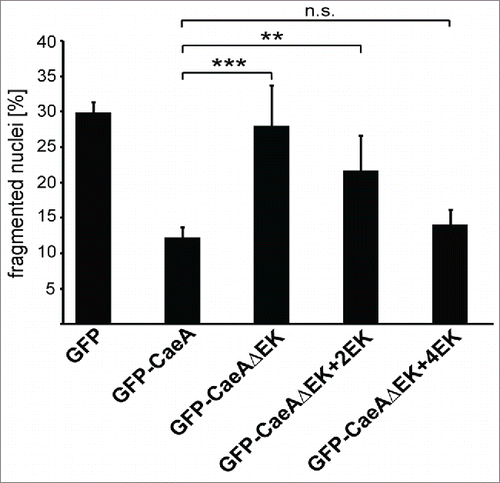
Table 1. Coxiella burnetii strains used in this study.
Table 2. Plasmids used in this study.
Table 3. PCR Primers used in this study.
Table 4. Sequencing Primers used in this study.
