Figures & data
Figure 1. Participants of the workshop under the porch of the Palace of Jabalquinto (Baeza, Spain). Photograph reproduced with permission of Joaquín Torreblanca López and the International University of Andalucia.

Figure 2. Selected molecular determinants of inter-bacterial interactions. Beneficial interactions include the exchange of material such as outer membrane components, metabolites or intracellular content and the transfer of plasmid DNA to recipient cells by conjugation. Biofilm formation and cell aggregation involve many determinants such as exopolysaccharides and Type IV pili, and allow an increased protection against antibiotics or physical stresses. Bacterial competition involves the release of antagonistic molecules, peptides and proteins or the direct delivery of toxin effectors by dedicated machineries, such as Type VI secretion and contact-dependent growth inhibition systems.
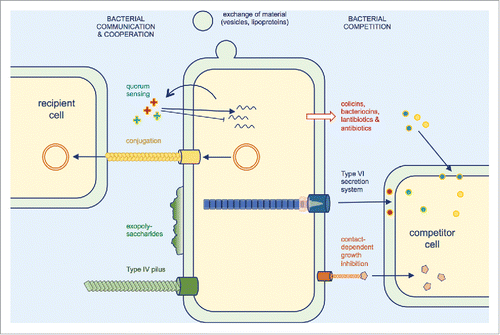
Figure 3. Selected adaptive mechanisms used by bacterial pathogens to survive within their hosts. These mechanisms include secretion and delivery of effectors which interfere with the host cytoskeleton and immune signaling. pH-, iron- and oxygen-dependent bacterial sensors can be activated to modulate expression of their regulons, leading to gene expression reprogramming and favoring bacterial adaptation. Dedicated bacterial enzymes can be used (i) to metabolize nutrients and (ii) to reduce or oxidize metabolites present in the host environment, both reactions conferring to the pathogen, an advantage over the competing microbiota.
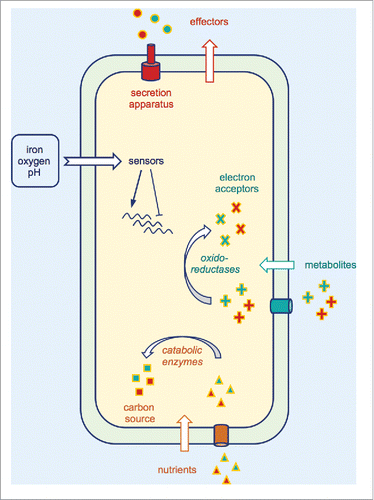
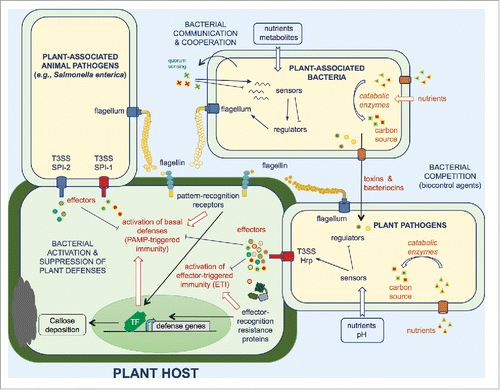
Figure 4. Selected adaptive mechanisms used by animal and plant pathogens as well as plant-associated bacteria to survive within their hosts. These mechanisms include secretion and delivery of effectors which interfere with host immune signaling. Bacterial sensors can be activated in response to different environmental cues such as pH or nutrients, to modulate expression of their regulons, leading to gene expression reprogramming and favoring bacterial adaptation. Dedicated bacterial enzymes can be used (i) to metabolize nutrients and (ii) to modify the host microenvironment, both reactions conferring to the pathogen an advantage over the competing microbiota. Biofilm formation and cell aggregation may also play a role in the adaptation to the plant environment and allow an increased protection against stresses. Bacterial competition involves the release of antagonistic molecules, peptides and proteins or the direct delivery of toxins.