Figures & data
Figure 1. Patterns of truncation of the C-terminal end of the NS1 protein in avian influenza viruses from 1902 to 2015. Shown are the truncations of the C-terminal end (ΔCTE) of NS1 among aa 202 to 230. Numbers on the left are total sequences pertaining the specific truncation form and between parentheses are the percentages of total ΔCTE divided by total 13026 analyzed AIV sequences. Truncated sequences are shown in pink while truncated sequences of CTE are in red. The naïve form of NS1, seen in 10582 out of 13026 sequences (81.2%) is 230 aa of length. Meanwhile, several truncations were observed in NS1 since 1954. The most common form is NS217 followed by NS224.
Table 1. Prevalence of NS1 truncation in the C-terminal end in 16 hemagglutinin subtypes of avian influenza viruses from 1902 to 2015.
Table 2. Prevalence of NS1 truncation in the C-terminal end in 9 neuraminidase subtypes of avian influenza viruses from 1902 to 2015.
Table 3. Prevalence of NS1 truncation in the C-terminal end among different HA-NA combination of avian influenza viruses from 1902 to 2015.
Table 4. Years and countries of origin of each form of NS1 C-terminal end truncation among avian influenza viruses from 1902–2015.
Figure 2. Prevalence of truncations in the C-terminus of NS1 of AIV among different bird species Blue columns represent the total sequences exhibiting truncation in the C-terminus of NS1 (NS1 ΔCTE) in AIV of a given species to the total AIV sequences with truncation (2331 analyzed in this study). Red columns refer to the total NS1 ΔCTE sequences to total sequences collected from the respective species. Results are shown in percentages (vertical axes).
Figure 3. In-vitro and in vivo characterization of recombinant HPAI H7N1 viruses generated in this study (a) Sequence of the NS1 protein of viruses generated in this study. (b) Replication kinetics in CEK cells after 2 independent trials after 1, 8, 24, 48 and 72 hours post infection. (c) Pathogenicity index after clinical examination of oculonasally infected 6-week-old specific-pathogen-free white leghorn chickens with 104.5 PFU/ml of the indicated viruses. Healthy chickens were scored “0.” Chickens showing one clinical sign (depression, ruffled feathers, diarrhea, sneezing, coughing, conjunctivitis, discharges, or cyanosis of comb, wattle or shanks) were scored “1” and were defined as ill. Severely ill chickens showing 2 or more clinical signs were scored “2,” whereas dead chickens were scored “3.” The pathogenicity index (PI) was calculated as the mean sum of the daily arithmetic mean values divided by 10; the number of observation days. (d) Virus excretion in oropharyngeal and cloacal swabs at 4 dpi was done by plaque assay and the mean titer ± standard deviation of positive birds was expressed in plaque forming unit per ml (PFU/ml).
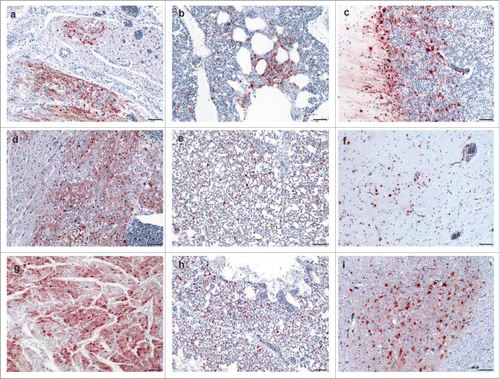
Figure 4. Detection of avian influenza virus nucleoprotein in infected chickens using immunohistochemistry Detection of influenza- nucleoprotein by immunohistochemistry (ABC method, intranuclear and intracytoplasmis bright red antigen signal by 3-amino-9-ethyl-carbazol chromogen and haematoxylin (blue) counterstain): heart (a, d, g), lung (b, e, h) and brain (c, f, i) samples obtained from birds infected with Hp-NS224 (a-c), HP- NS230 (d-f) and HP-NSLp (g-i) at 4 d post infection; for detailed data on the full tropism to different organs examined in this study readers are refered to Table S2. Bar = 50 µm.