Figures & data
Figure 1. The map and sources of water samples analyzed for detection of L. pneumophila in this study. Both the Kerman and Bam cities were located in south east of Iran. Number of hospitals, building (homes / hotels) and nursing houses that water samples were taken are included in this figure.
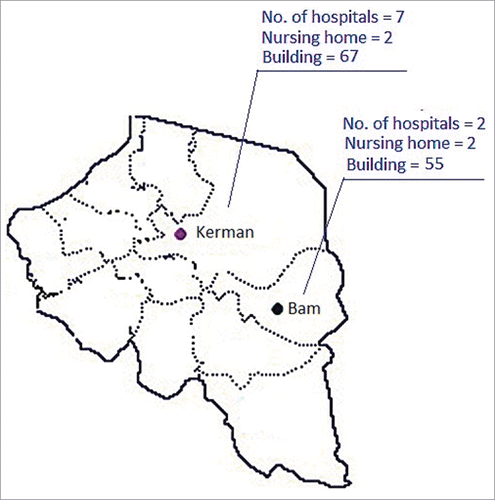
Figure 2. Agarose gel electrophoresis amplification of L. pneumophila by A) conventional-PCR (340bp) and B) nested –PCR (124bp) of the mip gene detected in cooling water samples in this study. Lanes 7-9 and 12 are water samples from Bam. Lane 11 is positive control (340bp). Lanes 1 and 14 are ladder consist of 100 base pairs DNA fragments. NC = negative control.
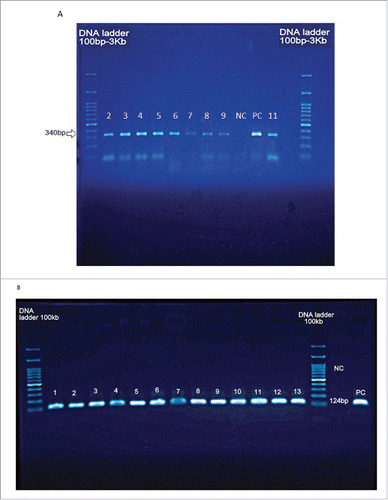
Figure 3. The real-time PCR detection of L. pneumophila mip gene isolated from water cooling systems investigated in this study. Panel: A) Real-time PCR of DNA extracted from water samples of Kerman and Bam. Panel: B) The standard curve with the CT plotted against the concentration of the starting quantity of template for each dilution
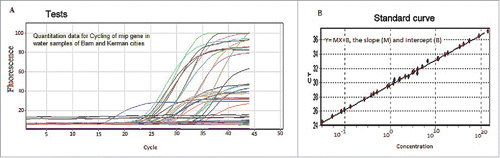
Figure 4. Dendrogram analysis of whole L. pneumophila genomic DNAs obtained from water samples of Bam and Kerman cities. Banding patterns were analyzed by UPGMA (unweighted pair-group method with arithmetic averages) clustering method using Gel Compare II software version 4.0 (Applied Maths, Sint-Matens-latem, Belgium). Degrees of homology were determined by Dice coefficient. Isolates that clustered >95% were considered related.
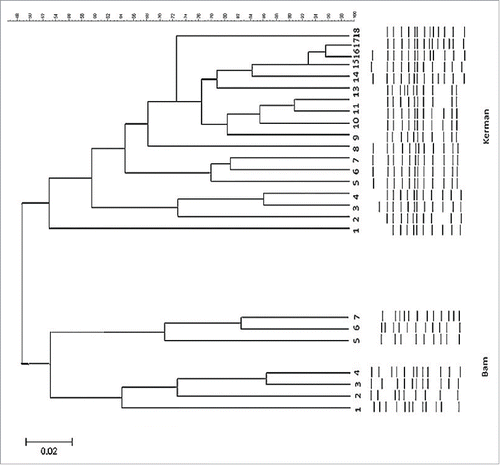
Table 1. Distribution of L. pneumophila mip gene detected in water samples collected from different cooling water systems in Kerman and Bam cities by nested and real-time PCR.
